Abstract
Nucleic acid-derived indices such as RNA/DNA ratios have been successfully applied as ecophysiological indicators to assess growth, nutritional condition and health status in marine organisms given that they provide a measure of tissue protein reserves, which is known to vary depending on changes in the environment. Yet, the use of these biochemical indices on highly mobile large predators is scarce. In this study, we tested the applicability of using nucleic acids to provide insights on the ecophysiological traits of two marine mammal species (common bottlenose dolphins and short-finned pilot whales) and explored potential related factors (species, sex, season, and residency pattern), using skin tissue (obtained from biopsy darts) of apparently healthy and adult free-ranging animals. Significantly higher RNA/DNA ratios were obtained for bottlenose dolphins (p < 0.001), and for visitor pilot whales when compared with resident pilot whales (p = 0.001). No significant changes were found between the sexes. Based on the percentile approach, the samples contain individuals in a general good condition (as the 10th percentile is not closer to the mean than the 75th percentile), suggesting that the studied region of Macaronesia may be considered an adequate habitat. The combination of this effective tool with genetic sexing and photographic-identification provided an overall picture of ecosystem health, and although with some limitations and still being a first approach, it has the applicability to be used in other top predators and ecosystems.
Similar content being viewed by others
Introduction
Understanding the physiology of an organism in function of its environment and the factors contributing to its variability can enable assessment of the relative impacts of anthropogenic and ecological pressures, which is essential in a changing world1,2. Among the most used ecophysiological indicators at the organism level in marine ecology are nucleic acid-derived indices, such as RNA/DNA ratios, RNA/mg and DNA/mg. Especially the former has been successfully applied as indicator of growth, nutritional condition and health status in marine organisms, as well as indicator of natural or anthropogenic impacts in marine populations and communities3. This is based on the fact that the concentration of cellular DNA is relatively constant in the somatic cells regardless of any changes in the organism’s environment, while the RNA content of a cell increases as the cellular demand for protein synthesis and growth increases4,5. It thus provides a measure of cellular protein synthesis capacity, which is generally interpreted as an indicator of tissue protein reserves, and that varies depending on changes in the environment where organisms live, such as food and habitat availability or physical factors6,7,8. Nevertheless, the use of these biochemical indices is unbalanced in favour to growth-related studies of microbial, invertebrate, fish and reptile communities9,10,11,12,13, in detriment to nutritional condition and health studies of highly mobile megafauna/apex predator species that may serve as potential bioindicators of the ecosystem.
Large-sized apex predators, such as cetaceans (marine mammals), play an important role in maintaining the structure and function of the environment they inhabit14,15, and are especially affected by increasing global anthropogenic pressures (e.g. direct and indirect catches, habitat destruction)16,17,18. Obtaining information into these species’ ecophysiological traits is thus important because of their conservation management. However, gathering physiological information from free-ranging cetaceans is challenging. Based on a commonly used technique of obtaining skin tissue at sea19, the present study tested the use of nucleic acid-derived indices to provide insights into the nutritional condition and health status of two cetacean species. To our best knowledge, this is the first study using RNA/DNA ratios as a bioindicator in cetaceans, and to a larger extent in wild mammals or highly mobile large predators.
The two cetacean species used in this study are the common bottlenose dolphin (Tursiops truncatus) and the short-finned pilot whale (Globicephala macrorhynchus). While the former is a mid-sized delphinid living in a fission-fusion society, is commonly found in coastal and oceanic temperate and tropical waters, and feeds on a large variety of bottom-dwellers and pelagic fishes and/or squids20,21, the latter is a large-sized delphinid living in a matrilineal society, occurs in oceanic habitats from tropical to warm-temperate waters, and feeds preferentially on squids caught during deep vertical dives22,23. On a global scale, both species have their conservation status described as least concern and their population trends are unknown24,25. In the north-east Atlantic, these species are considered common in the biogeographical region of Macaronesia26, which includes Madeira and the neighbouring archipelagos of the Azores and Canaries27,28,29. In this region, and for each species, animals with distinct residency patterns have been identified30,31,32,33, and genetic and photo-identification studies have shown inter-archipelago connectivity34,35,36,37. Yet, there is no information on these animals’ physiological condition to properly infer on their nutritional condition or health status, as neither on the influence of biological and environmental factors.
Taking advantage of the privileged location of Madeira to study the ecology of highly mobile large predators in a truly pelagic environment, our main goal was to test the applicability of using RNA/DNA ratios for assessing the ecophysiological condition in two cetacean species of different size and with distinct social structure and feeding habits, based on a multi-disciplinary approach. As specific goals, we explored (i) whether there are interspecific differences, and (ii) factors that may affect nutritional condition and health status at the intraspecific level, such as (a) sex, due to possible behavioural or hormonal differences, (b) season, due to possible environmental and/or gestation/lactating intra- and inter-annual differences, and (c) residency pattern, due to possible differences in the spatial structure and/or prey types.
Material and methods
Study site and data collection
Tissue samples were collected from free-ranging common bottlenose dolphins and short-finned pilot whales in the southern waters of Madeira Island (Portugal, Fig. 1). Madeira lies in a warm-temperate latitude, is surrounded by oceanic waters that are mainly oligotrophic, and is characterized by a narrow continental shelf, steep submarine canyons, and deep waters38,39. Field work comprised three dedicated campaigns covering distinct seasons: autumn 2017 (3rd to 20th of November), spring 2018 (16th of March to 21st of May), and autumn 2018 (28th of September to 05th of October). Exceptionally, three samples collected during late summer 2018 were included in the later bin in order to increase sample size (Fig. 2). Samples were obtained through a biopsy darting system (150-lb crossbow, with arrows and darts specially designed for small cetaceans by Finn Larsen, Ceta-Dart40), shot by experienced researchers carrying legal permits (see ‘Ethical approval’). Biopsy samples targeted the flanks of the animals, immediately below the dorsal fin. Samples were taken from apparently healthy adults, i.e. large and robust animals with no signs of emaciation41,42, and that were not carrying calves, i.e. not swimming in synchrony with young individuals. This minimized any potential bias associated with the analysis, given that the physiological condition of animals in poor health condition, calves, and lactating mothers might be different, and well as short-term stress induced by biopsying.
Location of Madeira and of the biopsied common bottlenose dolphins and short-finned pilot whales during 2017 and 2018 (map created with the software QGIS 2.18.15 https://www.qgis.org/en/site/). Species illustration by Les Gallagher - Fishpics & IMAR-DOP, UAç (the pilot whale is illustrated as a sub-adult in relation to an adult bottlenose dolphin).
Standardized RNA/DNA ratios of common bottlenose dolphins (n = 39) and short-finned pilot whales (n = 37) per sexes throughout the study period. Frequency histograms, means and percentiles 10, 25, 75, and 90th are used to illustrate a higher significant ecophysiological condition in bottlenose dolphins (p < 0.001, Table 2a); yet this difference should be interpreted with caution given that might be related to species and not to environmental conditions. The percentile approach shows that, in general, the lower percentile (10th) is not closer to the sample mean than the 75th percentile, which suggests that the sampled populations (especially of bottlenose dolphins) contain a high number of individuals in an adequate nutritional condition.
At sea, samples were immediately stored on liquid nitrogen. In the laboratory (LB3 of the University of Madeira), for each biopsy sample, the skin (0.5 cm in diameter) was separated from the blubber, and about ¼ of the skin tissue was separated for the determination of nucleic acids and another ¼ for the determination of sex, and stored at −80 °C. Prior to analyses, the skin tissue for the determination of nucleic acids was lyophilized for 48 hours at 60 °C and at a low pressure of about 10−1 atm, which was optimised for total removal of the water. Then, the skin tissue was weighed (±1 μg dry weight [DW]) on an electronic microbalance (Sartorius M5P).
Additionally, sighting data and individual identification photographs of the biopsied animals were taken. The sighting data comprised the coordinates (the closest to the individual or group), date, initial and end time, reaction to biopsy, species, presence of calves, age class, and best estimate of group size. Most parameters were determined at sea by experienced researchers and confirmed a posteriori using photographs. The collection of photographs from the cetaceans (left-and right-side of each individual) followed standard procedures43 using digital cameras with lenses and was simultaneous with biopsies.
Ecophysiological condition
Ecophysiological condition was assessed using the averages RNA (µg mg−1 DW), DNA (µg mg−1 DW), and sRD (standardized RNA/DNA ratios). The concentration of these nucleic acids was quantified following the procedures described in Caldarone et al.44 and Esteves et al.45. Briefly, the skin tissues of the biopsied cetaceans were chemically (cold sarcosyl Tris-EDTA extraction buffer) and mechanically (through sonification – 3 pulses 50 A for 1 min, and vortex) homogenised, and after centrifugation the tissues were isolated. Different volumes of supernatant were placed into fluorescent plate wells with Tris buffer according to sample dry-weight. Finally, 30 µl of specific nucleic acid fluorochrome dye GelRED (GR) was added into each well for the fluorescent reading of nucleic acids. Fluorescence was measured on a microplate reader (Biotek synergy HT model SIAFRTD) using an excitation wavelength of 365 nm and an emission wavelength of 590 nm, which allowed to determine the total stable amount of DNA and RNA (mainly ribosomal) in each sample. After the first read (total fluorescence of RNA and DNA), ribonuclease A (type-II A) solution was activated by incubating the Fluorescent Plates at 37 °C for 30 minutes. The plates were read again to determine the total amount of DNA fluorescence. In each plate where the samples were analysed “only-DNA” and “only-RNA” control sample were run, and a RNAase digestion was applied to all the samples and standards to be sure that RNA digestion was complete and no DNA degradation occurred.
Finally, RNA fluorescence was calculated as the difference between total fluorescence (first scan) and the fluorescence after RNAase activation (second scan). Standard curve of DNA-GR and RNA-GR with known concentrations of λ-bacteriophage DNA (0.25 μg μl−1) and 16S–23S E. coli RNA (4 μg μl−1) (Roche), were created to determine nucleic acids concentrations. The ratio of DNA and RNA slopes ranged from 4.74 to 7.10. RNA/DNA ratios were standardized based on this information and the reference slope ratio of 2.4 (following Caldarone et al.46). The determination of the nucleic acids was carried out at CCMAR (see affiliations) facilities.
Genetic sexing
Genomic DNA was extracted from the samples using a standard high-salt protocol as outlined in Sambrook et al.47. Multiplex PCR reactions aimed to amplify both ZFX and SRY gene fragments, as described in Rosel48, were carried out using Phusion Flash High-Fidelity PCR Master Mix (Thermo Scientific) in 20 µl reactions. The ZFX encodes a member of the krueppel C2H2-type zinc-finger protein family, while the SRY encodes a transcription factor that is a member of the high mobility group (HMG)-box family of DNA-binding proteins49 (Stelzer et al., 2016). The amplification conditions used in this study were as follows: initial denaturation for 10 seconds at 98 °C followed by 35 cycles of 1 second at 98 °C, 5 seconds at 51 °C, and a final extension of 15 seconds at 72 °C.
Due to difficulties in performing multiplex PCR reactions, we opted to perform single PCR reactions with only one set of primers (thus two PCR reactions per sample)48,50,51. To confirm whether the desired genes were amplified, several electrophoresis bands from different samples were sequenced. PCR products were cut from the gel, purified with the NZYGelpure (NZYTech) and sent to direct sequencing (Sanger sequencing) using the light run sequencing service of GATC Biotech (http://www.gatc-biotech.com/en/sanger-services/lightrun-sequencing.html). DNA sequences were analyzed using the BioEdit Sequence Alignment Editor version 7.0.4.152 and aligned against reference sequences from GenBank. The determination of the sex was carried out at CIIMAR (see affiliations) facilities.
Determination of residency patterns
Individual identification photographs of the biopsied pilot whales were compared with an existing long-term digital photographic-identification catalogue of this species. The catalogue comprises images collected in the same area where biopsies were taken, in the south of Madeira Island, since 2003. The catalogue was compiled by OOM (see affiliations) and most of its images were collected during whale-watching trips. The comparison was limited to pilot whales given that most bottlenose dolphins were not photographed properly during biopsying (move faster than pilot whales in general) and/or were not naturally well-marked. Of all the biopsied pilot whales, only one animal was not properly photographed (i.e. poor-quality photograph).
The compilation of the catalogue was based in Würsig and Würsig53, and detailed descriptions are given in Alves et al.35. Briefly, matching consisted of comparing the best processed image of each individual, based primarily on the number of unique notches on the dorsal fin, which allowed matching left-and right-side independently, and using fin shape or scars only to confirm matches54. Comparisons were carried out visually (e.g. Robbins et al.55), and only high-quality images (as illustrated in Alves et al.34) and matches with 100% certainty by three experienced researchers (F.A., M.W., A.D.) were used in the present study.
The establishment of residency patterns was based on the data set of individual-specific encounter histories (from 2003 to 2018). Individuals that exhibited multiyear and year-round site fidelity (photographed ≥15 times in at least three years and all four seasons; although most animals were photographed ≥63 times over 12 years) were termed residents; individuals photographed once (i.e. not previously catalogued) and not mixed with catalogued animals (to avoid erroneous classification of new/non-catalogued resident or visitor animals) were termed transients; and individuals that exhibited multi-year but seasonal specific presence (i.e. in only one or two seasons) were considered visitors (adapted from Alves et al.34). This was restricted to well-marked adult individuals in order to minimize erroneous classifications.
Data analyses
Descriptive (arithmetic means and percentiles) and inferential statistics (analysis of variance ‘ANOVA’ tests) were used to illustrate and test for significant differences (α = 0.05) in the biochemical indices between (a) species, (b) seasons (autumn 2017, spring 2018, and autumn 2018) and sexes in bottlenose dolphins, (c) seasons and sexes in pilot whales, and (d) residency patterns (residents, transients, and visitors) in pilot whales. The small sample size of some residency pattern bins did not allow us to test for differences between sexes and seasons within this variable. Significant differences were followed by a post hoc Tukey test. The Kolmogorov-Smirnov and Levene’s tests were used to test the ANOVA assumptions (normality and homogeneity of variances, respectively). All analyses were carried out using the R 3.5.3 statistical package56.
Results
Standardized RNA/DNA ratios (sRD) were obtained for 39 bottlenose dolphins (20 females and 19 males) and 37 pilot whales (8 females and 29 males) (Table 1). Ecophysiological condition index of bottlenose dolphins (mean = 0.49; percentiles 10, 25, 75, and 90th = 0.36, 0.40, 0.58, and 0.68, respectively) was significantly higher than pilot whales (mean = 0.37; percentiles = 0.23, 0.27, 0.44, and 0.56, respectively) (Table 2a, Fig. 2). The RNA and DNA concentrations were not significantly different between species (p = 0.40 for RNA, and p = 0.63 for DNA; SI1 and SI2, respectively).
In bottlenose dolphins, no significant changes were found in the sRD between sexes, between seasons, and neither in their interaction (Table 2b, Fig. 3). The RNA and DNA concentrations were significantly different in the factor season, with higher values in autumn 2018 (in both sexes) (SI1 and SI2). In pilot whales, the sRD was significantly different only between seasons (Table 2c.1), with higher values in autumn 2018 (in both sexes) (Figs. 3, SI3). The RNA and DNA concentrations showed the same pattern of the sRD, with significantly higher values in autumn 2018 (SI1 and SI2). The interaction of seasons and sexes could not be determined in pilot whales (i.e. one-way ANOVA tests were run independently for each factor instead of a two-way ANOVA) due to the low number of biopsied females in each season.
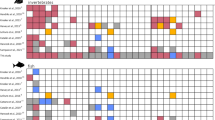
Biochemical condition (mean and percentiles 10 and 90th of the standardized RNA/DNA ratios) of bottlenose dolphins (a) and pilot whales (b) per sexes and seasons. Significant higher values (p < 0.001) were found only between autumn 2018 and the remaining seasons in pilot whales (Table 2c.1, SI3). The absence of significant differences between sexes (Table 2b for bottlenose dolphins and Table 2c.2 for pilot whales) increases the applicability of using RNA/DNA ratios in large vertebrates (due to reduced sex bias).
Residency patterns in pilot whales were attributed to 9 residents, 4 transients, and 17 visitors. The biochemical condition (sRD) was significantly higher in visitors (mean = 0.43; percentiles 10, 25, 75, and 90th = 0.27, 0.32, 0.52, and 0.56, respectively) than in residents (mean = 0.26; percentiles = 0.18, 0.25, 0.30, and 0.32, respectively) (Table 2d, Figs. 4, SI4). The RNA and DNA concentrations showed the same general pattern of the sRD (SI1 and SI2).
Standardized RNA/DNA ratios between pilot whales with distinct residency patterns throughout the study period. Frequency histograms, means and the percentile approach are used to illustrate a higher ecophysiological condition in visitors, which was significantly different from residents (p = 0.001, Table 2d, SI4). Based on the individual capture histories between 2003 and 2018, we classified: (i) residents, as individuals that exhibited multi-year and year-round (i.e. in the four seasons) site fidelity, (ii) transients, as individuals captured only once and not mixed with catalogued animals, and (iii) visitors, as individuals that exhibited multi-year but seasonal specific presence (i.e. in only one or two seasons); see ‘Material and methods’ for details.
Discussion
In this study, we used a percentile approach to obtain a better representation of the animal’s relative ecophysiological condition57. Because in field populations the condition is stochastically distributed, population sample percentiles can then describe the shape of the underlying distribution and the inter-individual variability, which facilitate the interpretation of what might be considered a poor or good condition57,58. In our case, and although with inter- and intra-specific differences (as discussed below), in general, the sample contains individuals in good condition given that the lower percentile (the 10th of RNA/DNA ratios - indicator of the lowest life-stage-specific condition57,59) is not close to the mean. Moreover, the 75th percentile is closer to the sample mean than the 10th percentile, which suggests an adequate health status. Hence, such relative nutritional condition of these highly mobile top predators indicate that the marine biogeographic region of Macaronesia may be considered an adequate habitat for those species, but that (especially in resident pilot whales) future population and ecosystem monitoring should be carried out (e.g. survival rates, PCB’s, heavy metals). Information on the ecophysiological condition of marine megafauna is scarce and mainly correlated with growth, and in the case of highly mobile predators such as cetaceans, most health indices have derived from photogrammetry of body condition or stress hormones60. In this study, and although preliminary, we demonstrate the use of biochemical indices as an innovative way to infer on the ecophysiological traits of bottlenose dolphins and pilot whales, and that these indices can provide an overall picture of ecosystem health.
Our specific goals explored differences in the ecophysiological condition at inter- and intra-specific levels, from which three main findings have emerged. These were based on the principle that individuals in good nutritional condition generally have high levels of RNA/DNA ratios, whereas individuals from related phylogenetic groups undergoing dietary restriction have a lower amount of RNA in their cells and hence a lower rate of RNA/DNA ratios1,3,61, given that the amount of RNA directly involved in protein synthesis varies with age, disease-state or environmental conditions62,63. Our first main finding is that RNA/DNA ratios varied significantly in animals with different residency patterns. RNA/DNA ratios were significantly higher in autumn 2018 in pilot whales, and significantly higher in visitors (when compared to resident pilot whales). But curiously, a closer inspection revealed that all pilot whales with a known residency pattern sampled in autumn 2018 were identified as visitors (Fig. 4). This suggests that the higher values obtained in autumn 2018 could correspond to visitors. This is supported by the higher RNA/DNA ratios from visitors (when compared to residents) in autumn 2017, and by the absence of significant differences in RNA/DNA ratios in bottlenose dolphins between seasons. Indeed, visitor pilot whales have different capture probabilities (when compared to those from residents and transients) in Madeira30,35, which should reflect a distinct biogeographic ecology for this ecotype, i.e. with different spatial structure, movements and/or feeding habitats. The higher RNA/DNA ratios thus suggest that visitor pilot whales are in better nutritional condition and are likely to be more robust or ‘adapted’ to a wider spectrum of pelagic habitats. Although different ecotypes displaying different feeding habits have been described in other cetacean species such as in killer whales (Orcinus orca) (e.g. Foote et al.64), there is no available information on their ecophysiological condition to support our findings. Therefore, the suggestion presented here should be viewed as a hypothesis to be explored in future research. This could include, for example, the assessment of the ecophysiological condition (based on the approached presented here), or the study of the nutritional ecology based on a geometry framework that distinguishes specific nutrients and calories65, in distinct ecotypes.
Second, no significant differences were obtained between sexes in any of the species. Chícharo et al.66 found higher RNA/DNA ratios in females than in males in fishes, crustaceans, and bivalves, suggesting that sexual dimorphism in addition to physiological and behavioural differences may have accounted for those differences. Therefore, those authors mentioned that the effect of sex should be taken into account on studies using nucleic acid concentrations. In this study, we used genetic sexing due to the difficulties in determining the sex of bottlenose dolphins and pilot whales at sea21,67, in order to comprise the effect of sex in our analyses. Taking into consideration that mammals lactate, which could imply different RNA for females, and that calves of both species have been commonly recorded in Madeiran waters27,68, higher variability in the RNA/DNA ratios could be expected in females, in at least one of the seasons. However, our findings showed similar ratios between sexes, which is in agreement with a study on smooth dogfish sharks (Mustelus canis)69. This suggests that, contrarily to invertebrates and small fishes, the RNA directly involved in protein synthesis does not vary significantly between females and males in large top predators; yet this lacks support from further studies. The fact that females and males do not provide different bias to the analysis could be a major advantage and applicability for the use of RNA/DNA ratios in large vertebrates.
Third, bottlenose dolphins showed significantly higher RNA/DNA ratios than pilot whales. However, and even if it is tempting to mention that this could be related to environmental conditions or reflect biological differences between the two species given all animals were biopsied close in time and space using the same in situ and laboratory methodological procedures, in fact we did not consider species-specific amounts of RNA and DNA and thus any differences might be related to species. Therefore, assuming a better nutritional condition in bottlenose dolphins (based for example on its more diversified diet20,21,70,71,72 or possible higher resilience to human-induced activities) should be interpreted with caution. In the future, such inter-specific discrepancy could be better interpreted if RNA/DNA ratios became available for other populations and/or species.
As an overview, this study has shown that RNA/DNA ratios can provide important information on the physiological and nutritional status of highly mobile large predators, supporting the use of these biochemical indices as indicators of the health status of marine organisms and ecosystems. Nevertheless, the application of this methodology on such animals offers limitations. First, obtaining tissue samples from free-ranging animals requires experienced researchers, and is expensive and time-consuming for most species; not to mention that many of these target taxa are endangered and/or not hunted commercially. Second, RNA/DNA ratios have been successfully used as important biomarkers of growth rates61,73, yet combining biochemical analyses with morphometric measurements is challenging in wild fast-moving megafauna due to difficulties in obtaining precise body lengths74. The same may apply to age class or sex determination (known only a posteriori in our case) of the sampled animal, which can be useful variables to take into account in data analysis. Additionally, different tissues or body parts can have different RNA or DNA-tissue relationships75, so caution should be made to obtain tissue samples from the same body part, as we did, in order to minimize potential bias. Finally, a comparison between studies or species requires similar metabolic responses and methodologies. Overall, the combination of methodologies used in this study (comprising biochemical analyses to determine the nucleic acid-derived indices, genetic analyses to determine sex, and photographic-identification to establish residency patterns), allowed a more comprehensive analysis of the ecophysiological condition in highly mobile large predators and in providing an ecological framework. Although this study constitutes a first approach in the use of nucleic acid derived indices in these animals, it deserves further attention to be used in other top predators and ecosystems.
Ethical approval
Biopsies were obtained in accordance with the relevant guidelines and regulations imposed by Instituto de Florestas e Conservação da Natureza (Instituto Português - Região Autónoma da Madeira) and under sampling permits 1.856/2017, 508/2018 and 10661/2018 from the same Portuguese institute.
References
Dahlhoff, E. P. Biochemical Indicators of Stress and Metabolism: Applications for Marine Ecological Studies. Annu. Rev. Physiol. 66, 183–207 (2004).
Gunderson, A. R., Armstrong, E. J. & Stillman, J. H. Multiple Stressors in a Changing World: The Need for an Improved Perspective on Physiological Responses to the Dynamic Marine Environment. Annu. Rev. Mar. Sci. 8, 357–378 (2016).
Chícharo, M. A. & Chícharo, L. RNA:DNA Ratio and Other Nucleic Acid Derived Indices in Marine Ecology. Int. J. Mol. Sci. 9, 1453–1471 (2008).
Ferron, A. & Leggett, W. C. An appraisal of condition measures for marine fish larvae. Adv. Mar. Biol. 30, 217–303 (1994).
Foley, C. J., Bradley, D. L. & Höök, T. O. A review and assessment of the potential use of RNA:DNA ratios to assess the condition of entrained fish larvae. Ecol. Ind. 60, 346–357 (2016).
Chícharo, L. et al. Diel variation of the RNA:DNA ratios in Crassostrea angulata (Lamarck) and Ruditapes decussatus (Linnaeus 1758) (Mollusca, Bivalvia). J. Exp. Mar. Biol. Ecol. 259, 121–129 (2001).
Holm-Hansen, O., Sutcliffe, W. H. Jr. & Sharp, J. Measurement of the deoxyribonucleic acid in the ocean and its ecological significance. Limnol. Oceanogr. 13, 507–514 (1968).
Lloret, J. & Planes, S. Condition, feeding and reproductive potential of white seabream Diplodus sargus as indicators of habitat quality and the effect of reserve protection in the northwestern Mediterranean. Mar. Ecol. Prog. Ser. 248, 197–208 (2003).
Dortch, Q., Roberts, T., Clayton, J. Jr. & Ahmed, S. RNA⁄DNA ratios and DNA concentrations as indicators of growth rate and biomass in planktonic marine organisms. Mar. Ecol. Prog. Ser. 13, 61–71 (1983).
Meesters, E., Nieuwland, G., Duineveld, G., Kok, A. & Bak, R. RNA⁄DNA ratios of scleractinian corals suggest acclimatisation⁄adapation in relation to light gradients and turbidity regimes. Mar. Ecol. Prog. Ser. 227, 223–239 (2002).
Mercaldo-Allen, R., Kuropat, C. & Caldarone, E. M. A model to estimate growth in young-of-the-year tautog, Tautoga onitis, based on RNA⁄DNA ratio and seawater temperature. J. Exp. Mar. Biol. Ecol. 329, 187–195 (2006).
Roark, A. M., Bjorndal, K. A., Bolten, A. B. & Leeuwenburgh, C. Biochemical indices as correlates of recent growth in juvenile green turtles (Chelonia mydas). J. Exp. Mar. Biol. Ecol. 376, 59–67 (2009).
Wagner, M., Durbin, E. & Buckley, L. RNA:DNA ratios as indicators of nutritional condition in the copepod Calanus finmarchicus. Mar. Ecol. Prog. Ser. 162, 173–181 (1998).
Bowen, W. D. Role of marine mammals in aquatic ecosystems. Mar. Ecol. Prog. Ser. 158, 267–274 (1997).
Roman, J. et al. Whales as marine ecosystem engineers. Front. Ecol. Environ. 12, 377–385 (2014).
Halpern, B. S. et al. A global map of human impact on marine ecosystems. Science 319, 948–952 (2008).
Jones, G. P., Srinivasan, M. & Almany, G. R. Population connectivity and conservation of marine biodiversity. Oceanography 20, 100–111 (2007).
Sousa, A. et al. How vulnerable are cetaceans to climate change? Developing and testing a new index. Ecol. Ind. 98, 9–18 (2019).
Noren, D. P. & Mocklin, J. A. Review of cetacean biopsy techniques: Factors contributing to successful sample collection and physiological and behavioral impacts. Mar. Mamm. Sci. 28, 154–199 (2012).
Connor, R., Wells, R., Mann, J. & Read, A. The bottlenose dolphin: social relationships in a fission-fusion society. In Mann, J., Connor R, R., Tyack, P. L. & Whitehead, H. (Eds). Cetacean societies: field studies of dolphins and whales. Chicago, University of Chicago Press, 91–126 (2000).
Wells, R. S. & Scott, M. D. Common bottlenose dolphin Tursiops truncatus. In Perrin, W. F., Würsig, B. and Thewissen, J. G. M. (Eds). Encyclopedia of marine mammals (2nd ed). Amsterdam, The Netherlands, Academic Press, 249–255 (2009).
de Soto, N. A. et al. Cheetahs of the deep sea: Deep foraging sprints in short-finned pilot whales off Tenerife (Canary Islands). J. Anim. Ecol. 77, 936–947 (2008).
Quick, N. J. et al. Hidden Markov models reveal complexity in the diving behaviour of short-finned pilot whales. Sci. Rep. 7, 45765 (2017).
Minton, G., Braulik, G. & Reeves, R. Globicephala macrorhynchus. The IUCN Red List of Threatened Species: e.T9249A50355227. https://doi.org/10.2305/IUCN.UK.2018-2.RLTS.T9249A50355227.en (2018).
Wells, R.S., Natoli, A. & Braulik, G. Tursiops truncatus. The IUCN Red List of Threatened Species: e.T22563A50377908. https://doi.org/10.2305/IUCN.UK.2019-1.RLTS.T22563A50377908.en (2019).
Spalding, M. D. et al. Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. BioScience 57, 573–583 (2007).
Alves, F. et al. Analysis of occurrence patterns and biological factors of cetaceans based on long-term and fine-scale data from platforms of opportunity: Madeira Island as a case study. Mar. Ecol. 39, e12499 (2018).
Carrillo, M., Pérez‐Vallazza, C. & Álvarez‐Vázquez, R. Cetacean diversity and distribution off Tenerife (Canary Islands). Mar. Biodivers. Rec. 3, 1–9 (2010).
Silva, M. A. et al. Spatial and temporal distribution of cetaceans in the mid‐Atlantic waters around the Azores. Mar. Biol. Res. 10, 123–137 (2014).
Alves, F. et al. Survival and abundance of short-finned pilot whales in the archipelago of Madeira, NE Atlantic. Mar. Mamm. Sci. 31, 106–121 (2015).
Dinis, A. et al. Bottlenose dolphin Tursiops truncatus group dynamics, site fidelity, residency and movement patterns in the Madeira Archipelago (North-East Atlantic). Afr. J. Mar. Sci. 38, 151–160 (2016).
Servidio, A. et al. Site fidelity and movement patterns of short‐finned pilot whales within the Canary Islands: Evidence for resident and transient populations. Aquat. Conserv. 29, 227–241 (2019).
Silva, M. A., Magalhães, S., Prieto, R., Santos, R. S. & Hammond, P. S. Estimating survival and abundance in a bottlenose dolphin population taking into account transience and temporary emigration. Mar. Ecol. Prog. Ser. 392, 263–276 (2009).
Alves, F. et al. Population structure of short-finned pilot whales in the oceanic archipelago of Madeira based on photo-identification and genetic analyses: implications for conservation. Aquat. Conserv. 23, 758–776 (2013).
Alves, F. et al. Complex biogeographical patterns support an ecological connectivity network of a large marine predator in the north-east Atlantic. Divers. Distrib. 25, 269–284 (2019).
Molina, C. et al. Large-scale movements of bottlenose dolphins (Tursiops truncatus) within the Macaronesia (NE Atlantic): dolphins with an international playground. Abstract book of the 32 nd Conference of the European Cetacean Society, La Spezia, Italy (2018).
Quérouil, S. et al. High gene flow in oceanic bottlenose dolphins (Tursiops truncatus) of the North Atlantic. Conserv. Genet. 8, 1405–1419 (2007).
Geldmacher, J., Van Den Bogaard, P., Hoernle, K. & Schmincke, H. U. The 40Ar/39Ar age dating of the Madeira Archipelago and hotspot track (eastern North Atlantic). Geochem. Geophy. Geosy. 1, 1999GC000018 (2000).
Martins, A. M. et al. Sea surface temperature (AVHRR, MODIS) and ocean colour (MODIS) seasonal and interannual variability in the Macaronesian islands of Azores, Madeira, and Canaries. Proc. SPIE 6743, Remote Sensing of the Ocean, Sea Ice, and Large Water Regions, 67430 A (2007).
Mathews, E. A., Keller, S. & Weiner, D. B. A method to collect and process skin biopsies for cell culture from the free-ranging gray whales (Eschrichtius robustus). Mar. Mamm. Sci. 4, 1–12 (1988).
Hart, L. B., Wells, R. S. & Schwacke, L. H. Reference ranges for body condition in wild bottlenose dolphins, Tursiops truncatus. Aquat. Biol. 18, 63–68 (2013).
Pettis, H. et al. Visual health assessment of North Atlantic right whales (Eubalaena glacialis) using photographs. Can. J. Zool. 82, 8–19 (2004).
Würsig, B. & Jefferson, T. A. Methods of photo-identification for small cetaceans. Rep. Int. Whal. Comm. 12, 42–43 (1990).
Caldarone, E. M., Wagner, M., St. Onge-Burns, J. & Buckley, L. J. Protocol and Guide for Estimating Nucleic Acids in Larval Fish Using a Fluorescence Microplate Reader. National Marine Fisheries Service, Woods Hole, MA. Available at http://www.nefsc.noaa.gov/nefsc/publications/crd/crd0111/crd0111.pdf (2001).
Esteves, E., Chícharo, M. A., Pina, T., Coelho, M. L. & Andrade, J. P. Comparison of RNA/DNA ratios obtained with two methods for nucleic acid quantification in gobiid larvae. J. Exp. Mar. Biol. Ecol. 245, 43–55 (2000).
Caldarone, E. M. et al. Intercalibration of four spectrofluorometric protocols for measuring RNA/DNA ratios in larval and juvenile fish. Limnol. Oceanogr.-Meth. 4, 153–163 (2006).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual. New York, Cold Harbor Spring Press (1989).
Rosel, P. E. PCR-based sex determination in Odontocete cetaceans. Conserv. Genet. 4, 647–649 (2003).
Stelzer, G. et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analysis. Curr. Protoc. Bioinformatics 54, 1.30.1–1.30.33 (2016).
Bérube, M. & Palsbøll, P. Identification of sex in cetaceans by multiplexing with three ZFX and ZFY specific primers. Mol. Ecol. 5, 283–287 (1996).
Richard, K. R., McCarrey, S. W. & Wright, J. M. DNA sequence from the SRY gene of the sperm whale (Physeter macrocephalus) for use in molecular sexing. Can. J. Zool. 72, 873–877 (1994).
Hall, T. A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows95/98/NT. Nucleic Acids Symp. Ser. 41, 9598 (1999).
Würsig, B. & Würsig, M. The photographic determination of group size, composition, and stability of coastal porpoises (Tursiops truncatus). Science 198, 755–756 (1977).
Urian, K. et al. Recommendations for photo-identification methods used in capture-recapture models with cetaceans. Mar. Mamm. Sci. 31, 298–321 (2015).
Robbins, J. et al. Return movement of a humpback whale between the Antarctic Peninsula and American Samoa: A seasonal migration record. Endanger. Species Res. 13, 117–121 (2011).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (2019).
Meyer, S. et al. On the edge of death: rates of decline and lower thresholds of biochemical condition in food-deprived fish larvae and juveniles. J. Mar. Syst. 93, 11–24 (2012).
Clemmesen, C. et al. Variability in condition and growth of Atlantic cod larvae and juveniles reared in mesocosms: environmental and maternal effects. J. Fish Biol. 62, 706–723 (2003).
Morais, P. et al. What are jellyfish really eating to support high ecophysiological condition? J. Plankton Res. 37, 1036–1041 (2015).
Hunt, K. E. et al. Overcoming the challenges of studying conservation physiology in large whales: a review of available methods. Conserv. Physiol. 1, cot006 (2013).
Vieira, S., Martins, S., Hawkes, L. A., Marco, A. & Teodósio, M. A. Biochemical indices and life traits of loggerhead turtles (Caretta caretta) from Cape Verde Islands. PLoS One 9, e112181 (2014).
Buckley, L. J., Caldarone, E. M. & Clemmesen, C. Multi-species larval fish growth model based on temperature and fluorometrically derived RNA/DNA ratios: results from a meta-analysis. Mar. Ecol. Prog. Ser. 371, 221–232 (2008).
Ikeda, T., Sano, F., Yamaguchi, A. & Matsuishi, T. RNA/DNA ratios of calanoid copepods from the epipelagic through abyssopelagic zones of the North Pacific Ocean. Aquat. Biol. 1, 99–108 (2007).
Foote, A. D. et al. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat. Commun. 7, 11693 (2016).
Machovsky-Capuska, G. E. & Raubenheimer, D. The Nutritional Ecology of Marine Apex Predators. Annu. Rev. Mar. Sci. 12, 15.1–15.27 (2020).
Chícharo, M. A., Chícharo, L., Amaral, A. & Morais, P. Sex effect on ratios and concentrations of DNA and RNA three in marine organisms. Mar. Ecol. Prog. Ser. 332, 241–245 (2007).
Augusto, J. F., Frasier, T. R. & Whitehead, H. Using photography to determine sex in pilot whales (Globicephala melas) is not possible: Males and females have similar dorsal fins. Mar. Mamm. Sci. 29, 213–220 (2013).
Reggente, M. A. L. et al. Nurturant behavior toward dead conspecifics in free‐ranging mammals: New records for odontocetes and a general review. J. Mamm. 97, 1428–1434 (2016).
Tavares, R., Lemus, M. & Chung, K. S. Evaluation of the instantaneous growth of juvenile smooth dogfish sharks (Mustelus canis) in their natural habitat, based on the RNA/DNA ratio. Cienc. Mar. 32, 297–302 (2006).
Fernández, R., Santos, M. B., Carrillo, M., Tejedor, M. & Pierce, G. J. Stomach contents of cetaceans stranded in the Canary Islands 1996-2006. J. Mar. Biol. Assoc. U.K. 89, 873–883 (2009).
Mintzer, V. J., Gannon, D. P., Barros, N. B. & Read, A. J. Stomach contents of mass stranded short-finned pilot whales (Globicephala macrorhynchus) from North Carolina. Mar. Mamm. Sci. 24, 290–302 (2008).
Olson, P. Pilot whales Globicephala melas and G. macrorhynchus. In Perrin, W. F., Würsig, B. & Thewissen, J. G. M. (Eds). Encyclopedia of marine mammals (2nd ed). Amsterdam, The Netherlands, Academic Press, 847–852 (2009).
Amaral, V., Cabral, H. N. & Paula, J. Implications of habitat specific growth and physiological condition for juvenile crab population structure. Mar. Freshwater Res. 59, 726–734 (2008).
van Aswegen, M. et al. Morphological differences between coastal bottlenose dolphin (Tursiops aduncus) populations identified using non-invasive stereo-laser photogrammetry. Sci. Rep. 9, 12235 (2019).
Houlihan, D. F., Hall, S. J., Gray, C. & Noble, B. S. Growth rates and protein turnover in Atlantic cod, Gadus morhua. Can. J. Fish Aquat. Sci. 45, 951–964 (1988).
Acknowledgements
To Ágatha Gil and Maria Paola Tomasino (CIIMAR) for helping with data collection, to Marisa Fernandes and Nereida Cordeiro (University of Madeira) for laboratory facilities, to Thomas Dellinger (University of Madeira) for logistical support, to Filipe Castro (CIIMAR) for helping with genetic analysis, to Tiago Marques (University of St Andrews) for helping with statistical analysis, to Gustavo Silva (OOM) for help with the creation of the map, to Les Gallagher for providing the species illustrations, and to the whale-watching operators Ventura, H2O-Madeira, VMT-Madeira and Lobosonda for contributing with photographic data. This study is a result of the project MARCET (MAC/1.1b/149) supported by MAC 2014–2020 programme under the Interreg fund, and of the project MarInfo (NORTE-01-0145-FEDER-000031) supported by NORTE 2020 under the PORTUGAL 2020 Partnership Agreement through the European Regional Development Fund (ERDF). This study had the support of the Oceanic Observatory of Madeira throughout the project M1420-01-0145-FEDER-000001-OOM, and of the Portuguese Foundation for Science and Technology (FCT) throughout the strategic projects UID/MAR/04292/2019 granted to MARE, UID/Multi/04326/2019 granted to CCMAR, and UIDB/04423/2020 & UIDP/04423/2020 granted to CIIMAR. F.A. and A.D. acknowledge Madeira’s Regional Agency for the Development of Research, Technology and Innovation (ARDITI) for funding their research throughout the project M1420-09-5369-FSE-000001.
Author information
Authors and Affiliations
Contributions
F.A. and M.A.T. conceived the study; F.A., A.D., E.D., I.S.P. and M.A.T. obtained funding for data collection and laboratory analyses; F.A., M.D., R.F., A.M.C., R.V., M.R. and A.D. collected the data; M.D., V.B. and M.A.T. determined the biochemical indices; R.V. and E.F. performed genetic sexing; F.A. and M.W. compiled the photo-identification catalogue/residency patterns; F.A. performed the statistical analysis; F.A. and M.D. drafted the manuscript; and all authors critically reviewed the manuscript and approved the version for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alves, F., Dromby, M., Baptista, V. et al. Ecophysiological traits of highly mobile large marine predators inferred from nucleic acid derived indices. Sci Rep 10, 4752 (2020). https://doi.org/10.1038/s41598-020-61769-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-61769-7
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.