Abstract
In this study, two chlorophyll A/B binding protein (CAB) genes (CsCP1 and CsCP2) in tea plant were cloned. The proteins encoded by these genes belong to the external or internal antenna proteins of PS II, respectively. They may be the targets of physiological regulation for tea leaf cell PS II because they all contain multiple functional domains and modifiable sites. The CAB gene family in the tea genome consists of 25 homologous genes. We measured the expression patterns of ten genes in the CsCP1 and CsCP2 subfamily under six different stresses. CsCP1 expression was inhibited in response to 6 kinds of stress; CsCP2 expression was slightly upregulated only after cold stress and ABA treatment. However, the expression levels of CSA016997 and CSA030476 were upregulated significantly in the six stresses. The results suggested that the 10 CAB genes may have different functions in tea leaves. Moreover, changes in the expression of the 10 genes under stress appear to be related to ABA- and MeJA-dependent signalling pathways, and their responses to MeJA treatment is faster than those to ABA. In addition, we introduced our experiences for cloning the genes in the context of complex genomes.
Similar content being viewed by others
Introduction
Chloroplasts are organelle involved in photosynthesis in plants and an important infrastructure to sustain the earth’s ecosystem. The light-harvesting complex (LHCs) consists of proteins and photosynthetic pigments and is an important functional components in chloroplasts. The main components of LHCs are CABs in higher plants. CABs are involved in light uptake, transmit the energy to the reaction centre of two photosystems (PS I and PS II), and adjust the distribution of the excitation energy between them and maintain the structure of thylakoid membrane1,2,3. CAB genes are exclusively encoded in the nucleus genome in higher plants, and are classified into 10 gene families based upon their nucleotide sequence homology. Four CAB gene families including LHCa1, LHCa2, LHCa3 and LHCa4, are associated with PS I, and the 6 CAB families related to PS II contain 3 major LHC II families (LHCb1, LHCb2 and LHCb3) and 3 minor LHC II families (LHCb4, LHCb5 and LHCb6, which encoded CP29, CP26 and CP24, respectively)4,5,6,7,8. Although the two photosystems in the thylakoid sub-domains of higher plants are comprised of different antenna proteins, the CAB proteins in PS II have attracted considerable attention gevin their complex physiological functions4,9. High homology exists between the major LHCs of PS II, which form homo- or heterotrimers to perform their functions. Compared with the major LHCII family proteins, the minor LHCII family proteins are integrated as monomer into the interior of PS II to absorb and deliver energy to the reaction centre3,10,11. CAB genes in some plants (including herbaceous and woody plant, such as Glycine max and Pyrus x bretschneideri) have been cloned. CAB gene expression is affected by light intensity, low temperature, high salinity, drought and disease11,12,13,14.
Evergreen woody plants in subtropical to temperate regions, such as tea plant (C. sinensis L.), have to adapt their photosynthetic capacity to the change greatly in light intensity and temperature during different seasons. Study on the genes encoding chloroplast proteins in evergreen woody plants and their expression patterns in various circumstances are important to understand the photosynthesis-mechanism changes in different seasons. There are some different mutant phenotypic cultivars related to the color of young leaves in C. sinensis, such as ‘Zijuan’ and ‘Baijiguan’. ‘Zijuan’ has a striking purple phenotype in young foliage and stems given an abnormal pattern of anthocyanin accumulation; ‘Baijiguan’ exhibits a yellow leaf phenotype, reduced chlorophyll (Chl) content, and aberrant chloroplast structures under high light intensity. Although Sun et al.15 and Wu et al.16 compared separately the transcriptome changes in ‘Zijuan’ or ‘Baijiguan’ with other tea cultivars’ during different light and temperature treatments, the relationship between tea phenotypes of the purple or yellow leaves and CAB genes were not mentioned in the two papers because the CAB genes were not cloned. Cheng et al.17 revealed that suppression of the P700-chlorophyll-a protein complex and LHCs might explain the albinism. Ma et al.18 found that LHCb in the Anji Baicha cultivar was suppressed during the albino stage, but increased dramatically when the shoots turned green. A proteomic analysis of tea leaves showed that CAB proteins were upregulated during drought stress19. However, little is known about the expression patterns of CAB genes and their gene families in C. sinensis. Here, two CAB genes in tea plants were cloned, and their characteristics, phylogenesis and gene family composition were analysed. This study can provide some information to further explain the functional characteristics and photosynthesis mechanism of CAB genes in tea plants.
Results
Cloning and characterization of two genes encoding the CAB of PSII in C. sinensis
In the analysis of wound induced transcriptome data (GEFQ00000000) from tea plants, a significantly downregulated CAB protein gene (comp56954_c0_seq1) was identified20. Then, we cloned the gene by RACE, and identified another homologous sequence in this process. Two genes encoding CAB proteins in tea plant were named CsCP1 and CsCP2 (KY709676 and KY709677) according to their molecular weights. The identity region is 943/949 (99%) between CsCP1 and comp59744_c0_seq1, a transcript from the wound-induced transcriptome in tea plant (GEFQ00000000), and no gap is observed. The different segments include shorted 15-bp and 10-bp nonsimilarity regions at the 5’ end and the loss of 210 bp at the 3′ end. The similarity region identity between CsCP1 and CSA004532 (972 bp), which was a gene from the C. sinensis genome database (http://www.plantkingdomgdb.com/tea_tree/), is 703/762 (92%), and a gap (30 bp) is noted. CsCP1 has an additional 30-bp fragment (yellow region). In addition, CsCP1 is 62 bp longer than CSA004532 at the 5′ end and 29 bp at the 3′ end (blue zone). Moreover, 99% (559/560) identity is noted between CsCP2 and comp56954_c0_seq1 (977 bp), but CsCP2 is 270 bp longer at the 5′ end and 417 bp shorter at the 3′ end. The sequence consistency is also 99% (793/798) between CsCP2 and CSA019572. However, CsCP2 is 14 bp longer at the 5 ‘end 18 bp longer at the 3′ end compared with CSA019572 (Fig. S1, Table S1).
The nucleotide sequences near CsCP1 and CsCP2 translation initiation codon (ATG) were all conformed to the Kozak rule (A/GNNAUGG)21 and contained the common TCAC/T sequences of the CAB family22. The ORF of the CsCP1 gene is 873 bp in length and encodes a 290-aa protein (30.96 kD, pI 6.33). Moreover, 33.10% a-helix, 37.59% random coil and 19.66% extended strand in the secondary structure of CsCP1 was predicted using SOPMA23. Using SMART analysis24, a typical CAB domain was composed of the 91- to 254-aa segment (164 aa in length) of the CsCP1 protein, which includes the binding sites for 4 chlorophyll-a, 3 chlorophyll-b and one 1,2-dipalmitoyl-phosphatidyl-glycerole. CsCP1 protein contains a SH3 (Src Homology-3) domain (146 to 207 aa) and two internal repeats in the 105 to 140 and 216 to 251 aa sections (the identity between two internal repeats is 44%) (Table S2). The ORF of CsCP2 gene is 798 bp in length, and encodes a 265-aa protein (28.65 kD, pI 5.97). The CsCP2 secondary structure contains 38.11% a-helix, 33.21% random coil and 17.36% extended strand. A typical CAB domain is also present in the 65 to 232 aa segment (168 aa in length) of CsCP2 protein, which includes the binding sites of 5 chlorophyll-a and one 1,2-dipalmitoyl-phosphatidyl-glycerole. CsCP2 protein contained a Rho domain (Rho subfamily of Ras-like small GTPases, 67 to 135 aa sections) and two internal repeats in the 73 to 122 and 189 to 237 aa sections (the identity between two internal repeats is 34%) (Table S2). Five predicted N-myristoylation sites are located in the 28–33, 32–37, 108–113, 122–127 and 178–183 aa segments of CsCP2 protein, and the 3 phosphorylation sites of protein kinase C are located in the threonines of 36–38, 40–42 and 234–236 aa segments of CsCP2. The nucleotide sequence alignment between CsCP1 and CsCP2 revealed two high similarity segments in a 222-bp 3′-terminal segment (the identity was 67%) and a 193-bp 5′-terminal segment (the identity was 70%) in the two genes, but the similarity of the other parts in their sequences was very low. The N-terminal regions of both proteins had a chloroplast transit peptide for chloroplast localization.
CsCP1 protein is the homologous protein of LHCb5 (KEGG orthology term K08916) as determined by KEGG pathway analysis, which suggested that it was the internal antenna protein of PS II. CsCP2 belongs to the external antenna protein of PS II because it is homologous to LHCb2 protein (KEGG orthology term K08913). Through SWISS-MODEL and Phyre2 analysis, the 214-aa residues of CsCP1 (74% of CsCP1 sequence) were modelled to cryoEM structure of spinach PSII-LHCII by the single highest scoring template with 100.0% confidence, and the 218-aa residues of CsCP2 (82% of CsCP2 sequence) were modelled to a Chlorophyll A/B binding protein. CsCP1 and CsCP2 had similar tertiary structures that all contained three a-helices that were integrated in the thylakoid membrane (Fig. 1). However, they have difference in binding pigment molecule types and numbers in other regions of the proteins except the CAB domain.
The CAB protein family constituents in C. sinensis and the phylogenetic relationship
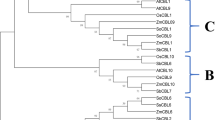
To identify protein homologous to CsCP1 and CsCP2 in tea plant, the genome database25 was searched by using the two CsCP1 and CsCP2 sequences as queries via Blastp with a threshold of E-value < 1e-5. A total of 25 homologous protein sequences were obtained after assessing for the presence of the Chloroa/b-bind domain in all obtained sequences. Phylogenetic analysis of the 25 sequences (MEGA6.0) indicated that they could be divided into 4 subfamilies, CsCP1 (CSA004532) and CsCP2 (CSA019572) were classified in the same subfamily, which contained 13 members (Fig. 2A, Table S3 and S4). There was only one amino acid difference between CsCP2 and the protein encoded by CSA019572.1 which was most similar sequence of CsCP2. However, there were 2 gaps (10 and 43 aa) between CsCP1 and its most similar sequence, the protein encoded by CSA004532.1, although the other sections were almost identical (Table S2).
Given the restriction of the tea tree genome assembly at present25,26, the genes encoding the 25 homologous proteins of CsCP1/ CsCP2 can not be localized to specific chromosomes in C. sinensis. However, the gene features of the abovementioned 25 genes are displayed by using GSDS2.0 (Gene Structure Display Server http://gsds.cbi.pku.edu.cn/) according to C. sinensis genome annotation25. Some of the genes did not contain introns, and others contained 1–6 introns (Fig. 2B). All of the 25 proteins encoded by the CAB gene family in tea plant contain a CAB domain, which consist of 2 or 3 α-helices. Seventeen of the genes have a chloroplast transport peptide in the N-terminus in addition to 8 additional sequences (Fig. 2C, Table S2).
To understand their evolutionary trajectories in the plant kingdom, proteins homologous to CsCP1 and CsCP2 were searched by using BlastP in NCBI. Greater than 100 homologous sequences of CsCP1 were obtained, including XP_012072637.1 (Jatropha curcas), XP_006478298.1 (Citrus sinensis), XP_002264295.1 (Vitis vinifera) and XP_010063732.1 (Eucalyptus grandis), all of which belonged to the chlorophyll A/B binding protein CP26. The sequence identities between CsCP1 and these proteins were greater than 76%, and the total score was greater than 440. Moreover, the sequence between CsCP1 protein and a hypothetical protein in Paenibacillus sp. IHB B 3415, WP_039310936, was completely identical. Greater than 100 CsCP2 homologous proteins were logged in NCBI, including XP_002531690.1 (a chlorophyll a-b binding protein 151, chloroplastic in Ricinus communis), XP_009802594.1 (a chlorophyll a-b binding protein 36 in Nicotiana sylvestris), XP_002271687.1 (chlorophyll a-b binding protein 151 in Vitis vinifera), XP_021664574.1 (a chlorophyll a-b binding protein 151 in Hevea brasiliensis). The sequence identities between CsCP2 and its homologous proteins were greater than 90%, and the total score were greater than 500. Sequence alignment was performed using 7 proteins selected from CsCP1 or CsCP2 homologous proteins, separately. The results indicated that most of the sequences of the two sets of proteins were highly similar, with the exception of their N-terminus regions and the regions near the second helix of the chlorophyll A/B binding domain (Fig. 3).
The expression profiles of the CAB genes in C. sinensis
To understand the expression patterns of the CAB gene family in tea plant, the expressions of 25 CAB genes after wounding was analysed firstly using the transcriptome data (GEFQ00000000)20. The result shows that the expression of almost all of the genes of the subfamily of CsCP1 and CsCP2 was significantly altered. The other three subfamily genes exhibited low expression with minimal changes (Fig. S2). Of the 13 genes in the CsCP1/CsCP2 subfamily, 3 genes (4th, 6th and 7th in Fig. 2: CSA035910, CSA035674 and CSA019509) lacked the coding region for the chloroplast transporter peptide. The expression levels of 6th and 7th genes were very low after wounding. The 11th gene in Fig. 2, CSA003567, is similar to 6th and 7th genes. The expression of these genes is very low, and their encoded proteins have only two transmembrane helix domains (Fig. 2; Table S2). Therefore, the three genes were not further analysed, and the expression patterns of other ten genes of the CsCP1 and CsCP2 subfamily in response to different stress conditions were assessed by qRT-PCR. The primers were designed in the differential region according to the sequence alignment of the CAB gene family in tea plants, and no nonspecific amplification occurred as verified by PCR (Fig. S3; Table S5). The main results are as follows: 1. During the 6 different stress treatments, the expression levels of only two of the 10 genes, CSA030476 and CSA016997, were upregulated transitorily greater than 1.5-fold compared with expression levels at 0 h′; Only CSA002361 was all upregulated momently in five stress conditions; CSA030474 and CSA008917 were upregulated in response to the three stressors. Three genes (CSA035910, CSA019572 and CSA016587) were upregulated by greater than 1.5-fold in two stress treatments. Of the upregulated 8 genes, 5 expressions were upregulated for longer than 3 h. In the other words, the time spans of the upregulated expressions were more than 2 serial sampling points, and the longest upregulated time was greater than 20 h (from 3 to 24 h sampling points). Six genes were upregulated transitorily by more than 4-fold in treatments, and the highest expression change was an 18011-fold increase compare with no treatment. However, two genes (CSA004532 and CSA024064) were all downregulated in response to six stressors. In addition, the expression levels of the abovementioned upregulated 8 genes were downregulated at the other sampling points, and their transcripts could not be detected at the most severe periods of downregulation (the expression level was 0). Some genes with closer genetic relationships to each other exhibit more similar expression patterns, for example, the expression patterns of CSA030474 and CSA030476, as well as CSA004532 and CSA024064 were similar. The expression patterns of CSA002361 and CSA016997 were also similar. However, the expression patterns of other genes were significantly different (Fig. 4; Table S6). 2. After mechanical injury, the expression levels of four (CSA030474, CSA030476, CSA002361 and CSA016997) of the 10 genes fluctuated between 0.02- and 12.82-fold that of the control expression levels. This result is different from that of the wound-induced transcriptome (GEFQ00000000), in which all of the genes were downregulated (Fig. S2 and S4A; Table S6). The expression levels of abovementioned 4 genes fluctuated between 0.01- and 4.2-fold that of the controls after NaCl treatment (Fig. S4D; Table S6). During the mannitol treatment, cold treatment, Methyl jasmonate (MeJA) treatment or abscisic acid (ABA) treatment, the expression levels of 4, 5, 5 and 7 genes were briefly upregulated by greater than 1.5-fold, respectively (Fig. S4C,B,F and E; Table S6). 3. The expression of CsCP1 gene was inhibited under the abovementioned six stress conditions, especially mannitol or NaCl treatment, and levels were reduced to less than 1/10 of the baseline value at some sampling points. CsCP2 gene expression was downregulated or not significantly changed in response to the four stress conditions, but was briefly upregulated to greater than 1.5-fold that of the control under cold and ABA treatment. Obviously, the expression pattern of CsCP1 is different from that of CsCP2. Moreover, it were also significantly different that the expression patterns of the genes between CsCP1 and CSA030476 or CSA016997, or between CsCP2 and CSA035910 or CSA008917, the coding proteins of which were with the highest similarity in amino acid sequences in the CAB gene family of C. sinensis (Table S7).
Discussion
Chloroplast has been always a research hotspot in the field of life sciences because it is a photosynthesis organelle in plant cells. However, it is also one of the most sensitive structures to multiple environmental stresses (e.g. cold, drought and wound) and plays a very important in plant stress response. Transcriptome analysis of chloroplast developmental defect mutants in Hordeum vulgare showed that approximately 67% of the cold regulatory genes were dependent on the organelle, and only 11% were chloroplast independent. The adaptation of plants to cold environment is achieved by changing the expression levels of genes that are closely related to the signal transduction from the chloroplast to cell nucleus27. Tea plants, as evergreen plant, are subjected to heat from strong light in summer and cold with excess light in winter, so their leaves represent an ideal material for studying photosynthesis mechanisms during cold/hot temperatures with strong light stress. To breed new variety of tea plants with early buds and freeze-resistant properties, our team mainly focused on the low temperature response in tea leaf and the chloroplast development28,29. Tea plants are also often subjected to picking and pruning injury given the economic significance of tea leaves. In this study, CsCP1 and CsCP2 genes were cloned according to wound-induced transcriptome data in tea plants20. It would be not easy to clone the corresponding genes using a RACE strategy based on the assembly sequences from a transcriptome due to the following reasons: 1. Some of the assembly sequences from the transcriptome data are not sufficiently accurate, especially given the absence of reference genome data or the lower quality of reference genome data25. 2. If a RACE strategy is used to clone a target gene of a polygenic family in a species, it is often interfered by the high similarity of nucleotide sequences among its members. 3. Multiple intron genes often produce some different cDNA sequences via selective splicing. Therefore, we experienced multiple failures in cloning the CsCP2 gene at the beginning of the study due to the abovementioned possible reasons. The two products, CsCP1 and CsCP2, were obtained though some optimization measures, such as analysing the gene family and re-designing the primers. It can be observed that the selection of primer locations is critical when we clone a gene according to the assembly sequence from a transcriptome or genome using RACE method, The primer would be avoided designing in the assembly regions where errors may occur and in the conserved sequence regions of the gene family, which is more important for qRT-PCR primer designs. There are two inferences that might be drawn from the abovementioned analysis and our results. 1. The sequence differences between CsCP1/CsCP2 and its corresponding assembled sequence based on the transcriptome or genome may result from the sequence is not assembled accurately (Fig. S1, Table S1). 2. The expression differences between CsCP1/CsCP2 and its corresponding assembled sequence may result also from the sequence is not assembled accurately (Figs. S2 and S4A, Table S6). Moreover, CsCP1 was completely identical to WP_039310936, a hypothetical protein from Paenibacillus sp. IHB B 3415. This finding represents an interesting problem given that a chlorophyll binding protein in tea plant has the same sequence as a protein in bacterial cells.
Plants are subjected to a variety of environmental stresses in their life cycles, and these stressors will directly or indirectly affect their photosynthesis processes. If CAB proteins play a rigid role, excess of light energy will be received because CAB proteins play an important role in the process of receiving and transmitting light energy. This situation will cause light damage to the photosynthetic apparatus of the chloroplast and even the entire mesophyll cell. Therefore, under environmental stress, CAB may represent a pivotal regulatory site of photosynthesis. Cells can regulate CAB activity at many levels, such as gene and protein levels. Structural analysis shows that CsCP1 protein contains a SH3 domain (Src Homology-3), and CsCP2 protein contains a Rho domain (Rho subfamily of Ras-like small GTPases), 3 protein kinase C phosphorylation sites and a phosphatase domain in its C-terminal (Table S2). These results suggest that the two proteins, especially CsCP2, can be used as targets for physiological regulation in the PS II of tea plant cells, and are affected by a variety of regulatory proteins. Furthermore, it suggests that phosphorylation/ dephosphorylation and GTP may be the main forms of the above regulation. The regulation of CAB protein levels enables chloroplasts to respond flexibly and rapidly to environmental stresses. In contrast, regulation at the genetic level has a lasting effect, although it is a delayed response. The CAB gene family in tea plant genome consisted of 25 members. There are 15, 9, and 15 CAB genes in the genome of Arabidopsis thaliana, rice or Populus, respectively30,31. Tea has more CAB genes than herbs (e.g., A. thaliana and rice) and deciduous woody plant (e.g., Populus), which may be related to the need for the leaves of evergreen woody plants to respond to more environmental stresses. In this study, the expression patterns of 10 genes in the CsCP1 and CsCP2 subfamily under six different stress conditions were detected by qRT-PCR to understand the role of CAB genes in tea plant. The expression levels of 8 genes were upregulated briefly by greater than 1.5-fold. The upregulation of CSA016997 and CSA030476 was the most significant among the 8 genes, and the upregulation was transitory or fluctuated in response to the six stresses. The upregulation of CSA016997 expression was the most rapid and significant for cold, mannitol and MeJA treatment. The response persisted from 3 to 6 or 12 h, and the expression level was upregulated by greater than 210-, 18011- or 2515-fold, respectively. The upregulation of CSA030476 was also very quick and significant in response to MeJA treatment, which was upregulated by 325- and 3516-fold compared with control at 3 to 6 h, respectively. The CSA002361 gene, which was the third gene most regulated, was upregulated under 5 stress treatments. However, regarding the 2 genes cloned in this study, CsCP2 gene expression was slightly upregulated only after cold stress and ABA treatment, while the expression level was downregulated at the other time points and the other four treatments. CsCP1 gene expression was inhibited in responsed to 6 types of stress (Fig. 4; Table S6). These findings may be directly related to the fact that CsCP1 and CsCP2 proteins are components of the pigment protein complex of the light-trapping antenna. Under stress conditions, reducing the synthesis of the light-trapping antenna proteins prevents the structure from receiving excessive light energy and thus reduces or avoids photooxidation11,32. According to this logic, the proteins encoded by the CSA016997, CSA030476 and CSA002361 genes, which were upregulated in response to stress conditions, should not be used to receive and transmit light energy, but rather to dissipate excess light energy or participate in photoprotection.
ABA and MeJA are all important phytohormones that modulate the plant respond to stressful conditions33,34,35. To explore the relationship between the expression regulation of CAB genes in tea plants and MeJA or ABA hormone signal transduction pathways under stress conditions, we analysed the expression changes in the CAB genes in tea leaves exposed to exogenous ABA or MeJA hormones. The results revealed that the expression levels of 7 and 5 genes were upregulated greater than 1.5-fold in response to ABA and MeJA treatments, respectively, compared with control. However, the expression of other genes was downregulated. These results suggest that the expression changes of the CAB genes in tea plant under stresses are related to ABA- and MeJA-dependent signalling pathways32,36,37. The upregulation of gene expressions were induced by MeJA occurs mostly at 3 or 6 h. The upregulation of gene expressions induced by ABA occurs mostly at 12 to 24 h, but a slight reduction occurred before the upregulation, suggesting that the responses of the CAB genes in the tea leaves to the MeJA is more rapid compared with that of ABA. However, the expression changes of analysed 10 CAB genes in tea plants in response to wound, cold, dehydration and high salt stress occurred from 3 h to 36 h. These expression changes and the upregulation timelines of some genes induced by the hormone and other stress treatments are closely related to each other, such as CSA016997. In contrast, the expressions of other genes exhibits minimal correlation with ABA and MeJA, such as CSA004532 and CSA024064 (Fig. S4; Table S6). These findings indicate: 1. the expression regulations of CABs in plant in response to stress may involve a variety of signal pathways, not just MeJA- and ABA-dependent pathway; 2. The functions of analysed 10 CAB genes in the CsCP1 and CsCP2 subfamily in tea plants may be more or less different, because their expression regulation in tea leaves in response to stresses are different.
In this study, two CAB genes in the tea plant were cloned, their gene family was analysed, and the expression patterns of ten genes in the CsCP1 and CsCP2 subfamily under six different stress conditions were measured by qRT-PCR. Our results provide some valuable data for further studies on photosynthesis and the function of CAB genes in tea plants.
Methods
Materials and treatments
Tea seedlings with consistent growth of 2-year-old clones from tea plant (C. sinensis L. cv. Xinyangdaye) were transplanted to pots in greenhouse. After 1 month of adaptation, the tea seedlings with consistent growth were moved into the artificial climate chamber (25 ± 1 °C; light/dark cycle, 14/10 h; light intensity, 500 μmol/m2/s; humidity, 75%) for 2 weeks. Then, the seedlings were treated and sampled.
For cold treatment, 10 tea seedlings were transferred to a growth chamber maintained at 2 °C.
For wounding treatment, each leaf of 20 plants was cut on both sides of the vertical midrib20.
For ABA/MeJA treatment, 20 plants were sprayed with ABA solution (100 μM)/ MeJA solution (45 µM), while the control plants were sprayed with water. The plants were maintained in the same growth chamber38,39.
The experimental materials used for dehydration stress were the branches of tea trees. The branches were washed with tap water immediately after being cut off to remove the wound hormones, immersed in 1/2 MS solution for one week to recover in the artificial climate chamber, and then transferred to mannitol solution (300 mM) for dehydration stress treatment38.
Each of the above treated materials were sampled at 0 (control check, CK), 3, 6, 12, 24 h and 36 h after the treatment. All of the first samplings (0 h) was conducted at 9:00 am.
All sampled materials were dipped immediately in liquid nitrogen and stored at a − 80 °C until use for RNA extraction.
Gene cloning and bioinformatic analysis of CAB sequences
Target genes were cloned from a tea plant cDNA library by RACE (rapid-amplification of cDNA ends) strategies using specific primers (Table S5) according to the previous experimental data of our laboratory. Homology search of CAB proteins was performed using BlastP (http://www.ncbi.nlm.nih.gov/BLAST/). ProtParam (http://www.expasy.org/tools) were used to compute the various physical and chemical parameters of CAB proteins. Protein structure analyses and domain identifications were enforced in SMART (http://smart.embl-heidelberg.de/), NCBI CDD40 (http://www.ncbi.nlm.nih.gov/cdd/), MOTIFSCAN (http://prosite.expasy.org/), Pfam41 (http://pfam.xfam.org/) and SWISS-MODEL (https://swissmodel.expasy.org/interactive) with default parameters to search for conserved domains or phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?d=index) in normal mode42. The transit peptides in proteins were predicted by ChloroP43 (http://www.cbs.dtu.dk/services/ChloroP/).
Analysis of CAB protein family members in C. sinensis and phylogenetic relationships
The C. sinensis genome database (http://www.plantkingdomgdb.com/tea_tree/ and http://www.yunchakj.com/index. aspx?lanmuid=63&sublanmuid=691&id=1367) was searched to identify the CAB domain-containing proteins using Blastp. Pfam and SMART were used to obtain protein sequences from the Chloroa/b-binding domain (PF00504, cl02879) using the Hidden Markov Model (HMM).
The sequences of CAB domain were used to identify conserved amino acid residues for interaction with co-factors. Multiple sequences alignments and regenerating phylogenetic tree were performed in Phylogeny.fr (http://www.phylogeny.fr/simple_phylogeny.cgi) using the Neighbour-Joining (NJ) method, and bootstrap analysis was conducted using 1000 replicates with the p-distance model.
Analysis of the expression profiles of the CsCP1 and CsCP2 gene family
qRT PCR analysis of CsCP1 and CsCP2 expression was performed using the ABI PRISM 7300 Real-Time PCR System (Applied Biosystems) according to the procedures by Wang et al.44. The qRT PCR primers (Table S2) were designed for differential sequence regions of the ten genes in the CsCAB family (Tables S3 and S4) using DNAClub software. The β-actin gene (HQ420251) of tea plant was used as reference for mRNA expression. Total RNA for qRT PCR was extracted from tea leaves via abovementioned treatments by using TaKaRa MiniBEST Plant RNA Extraction Kit (Code No: 9769). cDNA was synthesized from the total RNA (4 μg) using a PrimeScriptTM RT Master mix (Perfect Real Time) (TaKaRa Code No. RR036A).
References
Luciński, R. & Jackowski, G. The structure, functions and degradation of pigment binding proteins of photosystem II. Acta. Biochim. Pol. 53, 693–708 (2006).
Liu, C. et al. Structural and functional analysis of the antiparallel strands in the lumenal loop of the major light-harvesting chlorophyll a/b complex of photosystem II (LHCIIb) by site-directed mutagenesis. J. Biol. Chem. 283, 487–495 (2008).
Wei, X. et al. Structure of spinach photosystem II-LHCII supercomplex at 3.2Å resolution. Nature 534, 69–74 (2016).
Jansson, S., Stefánsson, H., Nyström, U., Gustafsson, P. & Albertsson, P. Antenna protein composition of PS I and PS II in thylakoid sub-domains. Biochimica et Biophysica Acta (BBA). Bioenergetics 1320, 297–309 (1997).
Green, B. R., Picheraky, E. & Kloppetech, K. Chlorophyll a/b binding proteins: an extended family. Trends Biochem. Sci. 16, 181–186 (1999).
Andersson, J. et al. Absence of the LHCb1 and LHCb2 proteins of the light-harvesting complex of photosystem II - effects on photosynthesis, grana stacking and fitness. Plant J. 35, 350–361 (2003).
Ifuku, K., Ishihara, S., Shimamoto, R., Ido, K. & Sato, F. Structure, function, and evolution of the PsbP protein family in higher plants. Photosynth. Res. 98, 427–437 (2008).
Plöchinger, M., Schwenkert, S., von Sydow, L., Schröder, W. P. & Meurer, J. Functional update of the auxiliary proteins PsbW, PsbY, HCF136, PsbN, TerC and ALB3 in maintenance and assembly of PSII. Front. Plant Sci. 7, 423 (2016).
Ido, K. et al. Cross-linking evidence for multiple interactions of the PsbP and PsbQ proteins in a higher plant photosystem II supercomplex. J. Biol. Chem. 289, 20150–20157 (2014).
Amarie, S. et al. Properties of zeaxanthin and its radical cation bound to the minor light-harvesting complexes CP24, CP26 and CP29. Biochim. Biophys. Acta. 1787, 747–752 (2009).
Xia, Y. et al. Allelic variations of a light harvesting chlorophyll A/B binding protein gene (LHCb1) associated with agronomic traits in barley. PLoS ONE 7, e37573 (2012).
Humbeck, K. & Krupinska, K. The abundance of minor chlorophyll a/b-binding proteins CP29 and LHCI of barley (Hordeum vulgare L.) during leaf senescence is controlled by light. J. Exp. Bot. 54, 375–383 (2003).
Caffarri, S., Frigerio, S., Olivieri, E., Righetti, P. G. & Bassi, R. Differential accumulation of LHCb gene products in thylakoid membranes of Zea mays plants grown under contrasting light and temperature conditions. Proteomics 5, 758–768 (2005).
Pavan, U. Genome-wide analysis of the family of light-harvesting chlorophyll a/b-binding proteins in Arabidopsis and rice. Plant Signal Behav. 5, 1537–1542 (2010).
Sun, B. et al. Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 6, 32534 (2016).
Wu, Q., Chen, Z., Sun, W., Deng, T. & Chen, M. De novo sequencing of the leaf transcriptome reveals complex light-Responsive regulatory networks in Camellia sinensis cv. Baijiguan. Front. Plant Sci. 7, 332 (2016).
Cheng, H., Chen, M., Yu, F. L. & Li, S. F. The variation of pigment-protein complexes in the albescent stage of tea. Plant Physiology Communications 36, 300–304 (2000).
Ma, C. L. et al. Differential expression analysis of different albescent stages of ‘Anji Baicha’ (Camellia sinensis L. O. Kuntze) using cDNA microarray. Sci. Hortic. 148, 246–254 (2012).
Wang, Y. et al. Proteomic analysis of Camellia sinensis (L.) reveals a synergistic network in the response to drought stress and recovery. J. Plant Physiol. 219, 91–99 (2017).
Li, X. et al. Transcriptome changes and its effect on physiological and metabolic processes in tea plant during mechanical damage. Forest Pathology e12432 (2018).
Kozak, M. Initiation of translation in prokaryotes and eukaryotes. Gene 234, 187–208 (1999).
Leutwiler, L. S., Meyerowitz, E. M. & Tobin, E. M. Structure and expression of three light-harvesting chlorophyll a/b-binding protein genes in Arabidopsis thaliana. Nucleic. Acids Res. 14, 4051–4064 (1986).
Geourjon, C. & Deleage, G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11, 681–684 (1995).
Letunic, I. & Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 4, D493–D496 (2018).
Xia, E. H. et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 10, 866–877 (2017).
Wei, C. et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 115, E4151–E4158 (2018).
Svensson, T. J. et al. Transcriptome analysis of cold acclimation in barley albina and xantha mutants. Plant Physiol. 141, 257–270 (2006).
Li, X. W. et al. A novel cold-regulated gene from Camellia sinensis, CsCOR1, enhances salt- and dehydration-tolerance in tobacco. Biochem. Biophys. Res. Commun. 394, 354–359 (2010).
Li, X. W., Liu, H. J., Xie, S. X. & Yuan, H. Y. Isolation and characterization of two genes of the early light-induced proteins of Camellia sinensis. Photosynthetica 2, 305–311 (2013).
Klimmek, F., Sjodin, A. & Noutsos, C. Abundantly and rarely expressed LHC protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 140, 793–804 (2006).
Zou, Z., Huang, Q. X. & An, F. Genome-wide identification, classification and expression analysis of LHC supergene family in castor bean (Ricinus communis L.). Agricultural Bitechnology 2, 44–48, 51 (2013).
Xu, Y. H. et al. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 63, 1095–1106 (2012).
Finkelstein, R. R., Gampala, S. S. L. & Rock, C. D. Abscisic Acid Signaling in Seeds and Seedlings. Plant Cell 14(Suppl), S15–45 (2002).
Adie, B., Chico, J. M., Rubio-Somoza, I. & Solano, R. Modulation of plant defenses by ethylene. J. Plant Growth Regul. 26, 160–177 (2007).
Lau, O. S. & Deng, X. W. Plant hormone signaling lightens up: Integretors of light and hormones. Curr. Opin. Plant Biol. 13, 571–577 (2010).
Baek, D. et al. A Role for Arabidopsis miR399f in Salt, Drought, and ABA Signaling. Mol. Cells 39, 111–118 (2016).
Gupta, A. et al. Global profiling of phytohormone dynamics during combined drought and pathogen stress in Arabidopsis thaliana reveals ABA and JA as major regulators. Sci. Rep. 7, 4017 (2017).
Tsugama, D., Liu, S. & Takano, T. Analysis of functions of VIP1 and its close homologs in osmosensory responses of Arabidopsis thaliana. PLoS One 9, e103930 (2014).
Król, P., Igielski, R., Pollmann, S. & Kępczyńska, E. Priming of seeds with methyl jasmonate induced resistance to hemi-biotroph Fusarium oxysporum f.sp. lycopersici in tomato via 12-oxo-phytodienoic acid, salicylic acid, and flavonol accumulation. J. Plant Physiol. 179, 122–32 (2015).
Marchler-Bauer, A. et al. CDD: NCBI’s conserved domain database. Nucleic. Acids Res. 43(Database issue), D222–226 (2015).
Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic. Acids Research 44, D279–D285 (2016).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Emanuelsson, O., Nielsen, H. & von Heijne, G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 (1999).
Wang, L. et al. Identification of genes induced in response to low-temperature treatment in tea leaves. Plant Mol. Biol. Rep. 27, 257–265 (2009).
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 31270727).
Author information
Authors and Affiliations
Contributions
X.W.L., Y.L.Z. and Z.J.G. conceived and designed the experimental plan, participated in sample collection and RNA preparation, analysed and interpreted the sequence data, drafted the manuscript and prepared the figures and tables. C.Y.C. and X.Y.L. participated in bioinformatics data analysis, and drafted and finalized the manuscript. T.T.Y., X.N.M. and F.D. participated in sample collection, RNA preparation and the PCR amplification of transcripts. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, XW., Zhu, YL., Chen, CY. et al. Cloning and characterization of two chlorophyll A/B binding protein genes and analysis of their gene family in Camellia sinensis. Sci Rep 10, 4602 (2020). https://doi.org/10.1038/s41598-020-61317-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-61317-3
This article is cited by
-
Genome-wide identification and expression profiling of photosystem II (PsbX) gene family in upland cotton (Gossypium hirsutum L)
Journal of Cotton Research (2024)
-
Fulvic Acid, Brassinolide, and Uniconazole Mediated Regulation of Morphological and Physiological Traits in Maize Seedlings Under Water Stress
Journal of Plant Growth Regulation (2023)
-
Analysis of Lhc family genes reveals development regulation and diurnal fluctuation expression patterns in Cyperus esculentus, a Cyperaceae plant
Planta (2023)
-
Genome wide identification and characterization of light-harvesting Chloro a/b binding (LHC) genes reveals their potential role in enhancing drought tolerance in Gossypium hirsutum
Journal of Cotton Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.