Abstract
In an attempt to find novel α-glucosidase inhibitors, an efficient, straightforward reaction to synthesize a library of fully substituted 6-amino-pyrazolo[1,5-a]pyrimidines 3 has been investigated. Heating a mixture of α-azidochalcones 1 and 3-aminopyrazoles 2 under the mild condition afforded desired compounds with a large substrate scope in good to excellent yields. All obtained products were evaluated as α-glucosidase inhibitors and exhibited excellent potency with IC50 values ranging from 15.2 ± 0.4 µM to 201.3 ± 4.2 µM. Among them, compound 3d was around 50-fold more potent than acarbose (IC50 = 750.0 ± 1.5 µM) as standard inhibitor. Regarding product structures, kinetic study and molecular docking were carried out for two of the most potent ones.
Similar content being viewed by others
Introduction
Recently, the world health organization (WHO) has identified the diabetes mellitus (DM) as a critical health challenge in the 21st century. The prevalence of diabetes has been increasing at alarming rate over the past three decades. This metabolic disorder, characterized by chronic hyperglycemia, leads to further severe damages like abnormally great thrust, excessive appetite, overweigh, blindness, excessive urination, cardiovascular complications, as well as renal and neurodegenerative diseases1,2,3,4,5,6. There are three main diabetes types among which type 2 or non-insulin dependent (T2DM) is the most common one, mainly treated by controlling the digestive enzyme activities such as α-glucosidase7,8,9.
α-Glucosidase, found in the brush-border surface membrane of intestinal cells, plays catalyzing role in the carbohydrate digestion process by which the postprandial blood glucose levels increases. Preventing the glucose release in the bloodstream, the α-glucosidase inhibitors control T2DM10. Additionally, this enzyme has a pivotal role in the biosynthesis of glycoprotein, therefore, its inhibitors have possessed anticancer, antitumor, antiviral, and immunoregulatory properties11,12,13,14,15. Acrabose, miglitol, voglibose, and deoxynojirimycin have clinically been used to restrict the α-glucosidase activity16. Considering the side effects and absorption problems associated with these drugs, new scaffolds should be synthesized and evaluated by medicinal chemists to extend the library of compounds17,18,19,20,21,22,23,24,25.
Pyrazoles are common structural motif in numerous drugs26 and biologically active compounds showing activities such as anti-cancer27,28, anti-inflammatory29, anti-hypertensive30, cannabinoid receptor antagonist31, dopaminergic receptor antagonist32, and α-glucosidase inhibitors33. Pyrazoles have been used to synthesize several other fused heterocycles. Pyrazolo[1,5-a]pyrimidines, in particular, have exhibited valuable pharmaceutical applications including various kinase inhibitors34,35,36,37, COX-2 inhibitors38, anti-viral (hepatetitis C and HIV)39,40, antimicrobial41,42,43, anxiolytic44,45, as well as positron emission tomography (PET) tumor imaging agents46. Some compounds containing this scaffold are approved and commercialized drugs, for example: Lorediplon A (for insomnia), Ocinaplon B (for anxiety), Zaleplon C and Indiplon D (for sedative and hypnotics), as well as Anagliptin E (for type 2 diabetes mellitus) Fig. 1)47. Furthermore, some pyrazolo[1,5-a]pyrimidines can be used in the treatment of diabetes48, obesity49, and CNS diseases50.
So far, different synthetic routes towards pyrazolo[1,5-a]pyrimidines have been reported. These methods have been mainly included the condensation of 3-aminopyrazoles with 1,3-bis electrophilic substrates51,52,53,54,55,56,57,58,59, 1,2-allenic lactones60, β-halovinyl aldehyde61, and activated alkynes62,63.
Through literature review, the reaction of α-azidochalcones 1 with 3-aminopyrazoles 2 to produce this scaffold has not proposed yet. Considering the significant mentioned roles in drug discovery process, synthesis of new derivatives of this scaffold is of increasing importance in medicinal organic chemistry. Therefore, we herein carried out an efficient, facile method to obtain highly substituted 6-amino-pyrazolo[1,5-a]pyrimidines 3 which have been studied to inhibit α-glucosidase.
Results
Chemistry
In this paper, we described a targeted reaction for the synthesis of a series of novel poly functionalized 6-amino-pyrazolo[1,5-a]pyrimidines 3 by Michael-addition-cyclization of α-azidochalcones 1 with 3-aminopyrazoles 2 (Scheme 1). It should be also noted that α-azidochalcones 1 have been applied over the last decade to prepare several valuable nitrogen containing skeletons64,65,66,67,68,69,70,71,72,73,74,75. To probe the generality of this strategy, various derivatives of both starting materials were applied under the appropriate reaction condition to afford a large library of corresponding 6-amino-pyrazolo[1,5-a]pyrimidines 3 in 65–92% yields.
A reasonable mechanism for this reaction is outlined in Scheme 2. Michael-addition of 3-aminopyrazole 2 from NH2 group to α-azidochalcones 1 and consequent removal of nitrogen molecule gives adduct 4. Next, an intramolecular nucleophilic attack of NH group in the pyrazole moiety to adjacent carbonyl group takes place to form pyrazolo[1,5-a]pyrimidine skeleton (intermediate 5). Finally, an imine-enamine tautomerization, followed by removal of a water molecule provides desired products 3.
In vitro α-glucosidase inhibitory activity
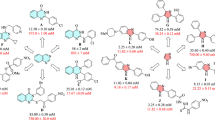
The obtained highly substituted 6-amino-pyrazolo[1,5-a]pyrimidines 3 were evaluated for their in vitro inhibitory activities against α-glucosidase (Saccharomyces cerevisiae, EC.3.2.1.20) and the results were compared with acarbose as the reference drug (Tables 1 and 2). As it can be seen, all the synthesized compounds showed good to excellent inhibitory activities with IC50 values of 15.2 ± 0.4−201.3 ± 4.2 µM in comparison to the standard drug IC50 = 750.0 ± 1.5 µM. To explain the structure and observed activity correlations, the 6-amino-pyrazolo[1,5-a]pyrimidines 3 were divided into two categories based on the substituents on the pyrazole moiety: the presence of amide functional group at C3-position 3a–z (summarized in Table 1) along with ester functional group at C3-position 3aa-ai (summarized in Table 2). Additionally, the substituents on the 5-phenyl and 7-phenyl rings of pyrimidine ring were changed in each series to optimize the α-glucosidase inhibition.
In the first category, the compounds3a–z were classified into three series according to N-aryl-pyrazole-3-carboxamide moiety: 1) unsubstituted derivatives 3a−i, 2) 4-methoxyphenyl derivatives 3j−r, 3) 4-chlorophenyl derivatives 3s−z.
Among the 6-amino-pyrazolo[1,5-a]pyrimidines 3a−i, compound 3d with 4-CH3 substitutent on the 5-aryl and 4-Br substituent on the 7-aryl ring showed the most inhibitory activity in this series (IC50 = 15.2 ± 0.4 µM). It is worth mentioning that this derivative showed the highest anti-α-glucosidase potency among all the synthesized compounds. Removal of the methyl group from 5-phenyl ring (compounds 3a and 3b) and also replacement of bromine with chlorine atom (compound 3c) led to the significant decrease in inhibitory activity. 4-OCH3 substituent on the 5-phenyl ring resulted into a considerable deterioration in activity (compounds 3e and 3f). Compound 3g with 4-Cl substituent on the 7-phenyl ring showed low activity (IC50 = 75.6 ± 5.0). Adding a chlorine atom to 5-position of phenyl ring, or replacing it with a heterocycle caused very good effect on the observed activities (compounds 3h and 3i).
In the second series, the 4-OCH3 substituted N-phenyl-pyrazolo-5-carboxamides 3j-r, compound 3m, which is the analog of compound 3d, showed the best activity against α-glucosidase. There was the same trend for the activities of compounds 3j-l with their analog in the first series. Compounds 3n and 3o with 4-OCH3 substituted 5-phenyl ring showed a moderate activity. The replacement of methoxy group with chlorine atom at 4-position of 5-phenyl ring led to the better performance (compounds 3p and 3q). Introducing a heterocycle on the 7-phenyl ring has a good effect to increase activity (compound 3r).
Among the synthesized derivatives in third series, compound 3v was found to be the most potent compound. Same as two previous series, removal of methyl group or addition of chlorine atom (compounds 3s-v) had a destructive effect on the observed inhibitory activities. It was found that introduction an electron-donating group (OCH3) on the 4-postion of 5-phenyl ring (compound 3w) causes a decrease in activity against α-glucosidase (IC50 = 53.7 ± 3.4). Finally, 4-Cl substituted 5-phenyl ring derivatives 3x-z were investigated. Compound 3x with unsubstituted 7-phenyl ring showed a weak inhibitory activity (IC50 = 80.3 ± 5.2). Introducing another chlorine atom to 4-position of this ring (compound 3y), or thiophene (compound 3z) led to a significant increase in inhibitory activity.
In the second category, the compound 3af with 4-OCH3 and 4-Cl substituents on the respectively 5 and 7-phenyl rings showed the highest potency against the α-glucosidase. Further changes on this compound like removing and replacing 4-OCH3 with 4-CH3 and 4-Cl on the 5-phenyl ring (compounds 3aa, 3ac and 3ah) as well as removing chlorine from 7-phenyl rings (compound 3ae) made notable increase in IC50 value. Compound 3ag with 4-Cl on the 5-phenyl ring was the weakest compound in this series. Addition of another chlorine atom to 4-position of this ring (compound 3ah), or thiophene (compound 3ai) improved the inhibition activities.
Thorough the comparison of IC50 values of synthesized3a–z with their analog 3aa-ai, it can be found that substituents on the pyrazole moiety played a substantial role on the observed α-glucosidase inhibitory activities. Although the presence of 4-OCH3 on the 5-phenyl ring had destructive effect in the first category, the compounds containing this group showed the highest activities in the second category.
Enzyme kinetic studies
The inhibition mode of the synthesized compounds 3 against α-glucosidase was investigated. For this purpose, kinetics analysis was carried out with reference drug, acarbose, and the most potent derivative in each category (3d and 3af). The inhibition type was indicated on the basis of Michaelis-Menten and Lineweaver-Burk plots. As it can be seen in the Lineweaver-Burk plot of selected compounds (Fig. 2), with increasing inhibitor concentrations, the Km value gradually increased while Vmax value remained unchanged which indicated competitive inhibition. Accordingly, this study revealed both 3d and 3af compete with acarbose for binding to the enzyme active site. Furthermore, plot of the Km versus different concentration of inhibitor gave an estimate of the inhibition constant, Ki of 12 µM and 65 µM for compounds 3d and 3af, respectively.
Enzyme Kinetic Studies: (a) The Lineweaver–Burk plot in the absence and presence of different concentrations of compound 3d. (b) The secondary plot between Km and various concentrations of compound 3d. (c) The Lineweaver–Burk plot in the absence and presence of different concentrations of compound 3af. (d) The secondary plot between Km and various concentrations of compound 3af.
In-silico ADME evaluation
The ADME properties for some of the synthesized highly substituted 6-amino-pyrazolo[1,5-a]pyrimidines 3 were computed using Swiss ADME online (http://www.swissadme.ch/index.php) toolkit76. Through this in-Silico study under the Lipniski’s rule, five determined drug-likeness parameters were compared with the known drugs77. These evaluated parameters are summarized in Table 3. On the basis of MW (<500), HBA (≤10), HBD (<5), and log P (<5) values, the good oral bioavailability of the selected compounds can be estimated. Lipophilicity is determined by Log P in which P is the octanol-water partition coefficient. As it can be seen in Table 3, all the studied compounds have the Log P values in the desirable range. The molecular flexibility can be proved regarding the number of rotatable bonds which should be less than 10 (nROTB <10) and regarding to Table 3, all the obtained numbers are 6 and 7. Topological polar surface area (TPSA) can reveal the surface contribution of polar fragments. The high value of TPSA (>140 Å2) may show low blood-brain barrier (BBB) penetration, and therefore, poor membrane permeability78. As it can be seen in Table 3, the TPSA values of the tested compounds are in the range 82.51–110.75 Å2 exhibiting their permeability in the cellular plasma membrane. The total number of hydrogen bond donors (HBD) should be <5, and the total number of hydrogen bond acceptors (HBA) should be ≤10. All compounds reveal HBDs of 1 and HBAs of 6 and 7 values. Additionally, according to the Veber rule79, the number of rotatable bonds should be ≤10 and TPSA <140 Å2, or sum of HBD and HBA<12, therefore, all compounds were shown to have good oral bioavailability. Finally, in consistent with ADME predictions, all the studied compounds were proved to have positive drug-likeness values.
Cytotoxicity studies
The cytotoxicity of some of potent compounds including 3d, 3m, 3v, and 3af was evaluated through use of the breast cancer cell line MDA-MB-231 and human pancreatic cancer cell line PANC-1. The results proved that at concentration of 100 µM, these selected compounds did not possess any cytotoxic activity against the mentioned cell lines (IC50 > 200 µM).
Molecular docking studies
Since there was not any X-ray crystallographic structure of the Saccharomyces cerevisiae α-glucosidase in the RCSB protein data bank, a homology modeling method was performed by Auto Dock Tools (version 1.5.6) to study ligand-enzyme interactions21,80. Briefly, crystal structures of isomaltase from Saccharomyces cerevisiae (PDB code 3A4A), with 72% identical and shares 85% similarity with the Saccharomyces cerevisiae α-glucosidase, was designated for building modeled α-glucosidase. Afterward, the interaction modes of acarbose as standard inhibitor and the most potent compound in each category 3d and 3af in the active site of α-glucosidase were studied.
As shown in Fig. 3, acarbose formed interactions with Asn241, His279, Glu304, Arg312, Thr302, Thr307, Ser308, and Gln322 residues in the enzyme active site. For the most active compound 3d, amino group established hydrogen bonds with active site residues Thr307 and Glu304. Furthermore, 4-bromo phenyl moiety formed a π-anion interaction with Glu304. Several hydrophobic interactions were also observed with the active site residues His239, Pro309, Arg312 and Ala326. The interactions of 3d are shown in Fig. 4a. In the case of 3af (Fig. 4b), hydrogen bonds between amino group and the active site residues Thr307 and Glu304 were formed. Glu304 interacted with 4-choloro phenyl moiety to form π-anion interaction. In addition, several hydrophobic interactions were observed between His239, Val305, Pro309, and Arg312 and 4-choloro phenyl moiety. Further studies on binding energies of compounds 3d, 3af and acarbose revealed that they have lower free binding energy (3d: −10.0 kcal/mol and 3af: −9.57 kcal/mol) than acarbose (−4.04 kcal/mol). This means they can bond easier to the target enzyme in comparison to acarbose.
Conclusion
In conclusion, we have represented a Michael-addition-cyclocondensation reaction between α-azidochalcones and 3-aminopyrazoles to prepare a novel library of fully substituted pyrazolo[1,5-a]pyrimidines and evaluated their α-glucosidase activities. Providing an efficient, simple protocol from readily available starting materials, this method led to new 6-amino-pyrazolo[1,5-a]pyrimidines in short time and under the mild conditions. Additionally, easy work-up without any need for chromatography purification processes and really good product yields are the significant features of this proposed reaction. The synthesized compounds were investigated by α-glucosidase inhibitory activity assay. All of them showed very good to excellent activities in comparison to the standard drug. Among these derivatives, 3d was the most potent one with IC50 value of 15.2±0.4 µM. The kinetic analysis for the most active compound from each category (3d and 3af) compound showed that there was a competitive mechanism to inhibit α-glucosidase. Furthermore, docking studies for these products revealed there were several interactions between desired compounds and important amino acids in the active site of the enzyme.
Experimental
Methods
All chemicals were purchased from Merck (Germany) and were used without further purification. Melting points were measured on an Electrothermal 9100 apparatus. Elemental analyses for C, H and N were performed using a Heraeus CHN-O-Rapid analyzer. Mass spectra were recorded on an Agilent Technologies (HP) 5973 mass spectrometer operating at an ionization potential of 20 eV. IR spectra were recorded on a Shimadzu IR-460 spectrometer. 1H and 13C NMR spectra were measured (in chloroform (CDCl3) and dime-thyl sulfoxide (DMSO-d6) solutions) with Bruker DRX-500 AVANCE (at 500.1 and 125.8 MHz) instruments. α-azidochalcones 1 as well as 3-amino-N-aryl-5-(phenylamino)-1H-pyrazole-4-carboxamides 2a–c and ethyl 3-amino-5-phenyl-1H-pyrazole-4-carboxylate 2d were obtained from synthetic methods reported in the literature81,82,83.
General Procedure for the Synthesis of Highly Substituted 6-Amino-N,5,7-Triaryl-2-(Phenylamino)Pyrazolo[1,5-a]Pyrimidine-3-Carboxamide (3a-z)
A solution of α-azidochalcone 1 (1 mmol), 3-amino-N-aryl-5-(phenylamino)-1H-pyrazole-4-carboxamide 2a–c (1.5 mmol), and sodium hydroxide (NaOH) (1.5 mmol) in N,N-dimethyl formaldehyde DMF (5 mL) was stirred at 80 °C for 10 min. After completion of the reaction according to TLC analysis, the obtained mixture was cooled down to ambient temperature. Then, water was added (10 mL) and extracted three times with ethylacetate (EtOAc) (15 mL for each time). The combined organic extracts were washed with brine, dried over Na2SO4, and then concentrated. The precipitated product was filtered and recrystallized in 5:1 n-Hexane/EtOAc to afford pure compound 3a–z.
6-Amino-7-(4-chlorophenyl)-N,5-diphenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3a)
Dark yellow solid; yield: 86%, mp 248–249 °C. IR (KBr) (νmax/cm−1): 3453–3236 (2NH and NH2), 1649 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.85 (s, 1H, amide NH), 9.26 (s, 1H, NH), 8.07 (d, J = 7.6 Hz, 2H, 2CH), 7.82 (d, J = 7.6 Hz, 2H, 2CH), 7.72–7.66 (m, 5H, 5CH), 7.62 (d, J = 7.7 Hz, 2H, 2CH), 7.51 (d, J = 7.4 Hz, 2H, 2CH), 7.36 (t, J = 7.3 Hz, 2H, 2CH), 7.20 (t, J = 7.2 Hz, 2H, 2CH), 7.08 (t, J = 7.4 Hz, 1H, CH), 6.88 (t, J = 7.3 Hz, 1H, CH), 4.51 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.2, 148.8, 140.1, 139.7, 138.2, 135.1, 134.1 and 130.6 (8C), 130.0 (2CH), 129.9 (CH), 129.5 (2CH), 128.5 (2CH), 128.41 (2CH), 128.37 (2CH), 128.30 (2CH), 127.9 and 127.5 (2C), 122.6 and 120.2 (2CH), 118.3 (2CH), 116.4 (2CH), 85.2 (C). EI-MS, m/z (%): 533 (M+ 37Cl, 27), 531 (M+ 35Cl, 75), 438 (100), 413 (13), 307 (8), 265 (15), 244 (7), 203 (8), 151 (9), 138 (13), 117 (12), 104 (21), 93 (96), 77 (53), 66 (57), 51 (10). Anal. Calcd for C31H23ClN6O (531.02): C, 70.12; H, 4.37; N, 15.83. Found: C, 70.19; H, 4.43; N, 15.78%.
6-Amino-7-(4-bromophenyl)-N,5-diphenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3b)
Dark yellow solid; yield: 79%, mp 235–237 °C. IR (KBr) (νmax/cm−1): 3436–3258 (2NH and NH2), 1656 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.83 (s, 1H, amide NH), 9.25 (s, 1H, NH), 7.99 (d, J = 7.3 Hz, 2H, 2CH), 7.88–7.77 (m, 4H, 4CH), 7.73–7.64 (m, 3H, 3CH), 7.62 (d, J = 7.1 Hz, 2H, 2CH), 7.50 (d, J = 7.3 Hz, 2H, 2CH), 7.37 (t, J = 7.3 Hz, 2H, 2CH), 7.20 (t, J = 7.0 Hz, 2H, 2CH), 7.08 (t, J = 7.4 Hz, 2H, 2CH), 6.88 (t, J = 7.2 Hz, 2H, 2CH), 4.48 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.2, 148.8, 140.1, 139.7, 138.2 and 135.4 (6C), 131.3 (2CH), 130.6 (C), 130.2 (2CH), 129.9 (CH), 129.5 (2CH), 128.5 (2CH), 128.4 (2CH), 128.3 (2CH), 127.8, 127.5 and 123.0 (3C), 122.6 and 120.2 (2CH), 118.3 (2CH), 116.4 (2CH), 85.2 (C). Anal. Calcd for C31H23BrN6O (575.47): C, 64.70; H, 4.03; N, 14.60. Found: C, 64.79; H, 3.92; N, 14.72%.
6-Amino-7-(4-chlorophenyl)-N-phenyl-2-(phenylamino)-5-p-tolylpyrazolo[1,5-a]pyrimidine-3-carboxamide (3c)
Dark yellow solid; yield: 82%, mp 232–233 °C. IR (KBr) (νmax/cm−1): 3465–3278 (2NH and NH2), 1652 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.86 (s, 1H, amide NH), 9.26 (s, 1H, NH), 8.06 (d, J = 7.8 Hz, 2H, 2CH), 7.73 (d, J = 7.3 Hz, 2H, 2CH), 7.70 (d, J = 7.9 Hz, 2H, 2CH), 7.62 (d, J = 7.3 Hz, 2H, 2CH), 7.53 (d, J = 7.8 Hz, 2H, 2CH), 7.50 (d, J = 7.9 Hz, 2H, 2CH), 7.37 (t, J = 7.2 Hz, 2H, 2CH), 7.22 (t, J = 7.3 Hz, 2H, 2CH), 7.08 (t, J = 7.2 Hz, 1H, CH), 6.89 (t, J = 7.0 Hz, 1H, CH), 4.47 (s, 2H, NH2), 2.48 (s, 3H, CH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.2, 148.7, 140.2, 139.8, 139.7, 138.2, 135.1, 134.1 and 130.8 (9C), 130.0 (2CH), 129.4 (2CH), 128.9 (2CH), 128.5 (2CH), 128.4 (2CH), 128.3 (2CH), 127.9 and 124.5 (2C), 122.6 and 120.2 (2CH), 118.3 (2CH), 116.5 (2CH), 85.2 (C), 20.7 (CH3). EI-MS, m/z (%): 547 (M+ 37Cl, 17), 545 (M+ 35Cl, 49), 530 (12), 452 (96), 438 (27), 395 (23), 293 (56), 200 (93), 93 (100), 77 (85), 65 (66), 55 (39), 43 (51). Anal. Calcd for C32H25ClN6O (545.04): C, 70.52; H, 4.62; N, 15.42. Found: C, 70.48; H, 4.56; N, 15.37%.
6-Amino-7-(4-bromophenyl)-N-phenyl-2-(phenylamino)-5-p-tolylpyrazolo[1,5-a]pyrimidine-3-carboxamide (3d)
Dark yellow solid; yield: 78%, mp 271–272 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.86 (s, 1H, amide NH), 9.26 (s, 1H, NH), 7.98 (d, J = 7.6 Hz, 2H, 2CH), 7.83 (d, J = 7.4 Hz, 2H, 2CH), 7.72 (d, J = 7.2 Hz, 2H, 2CH), 7.61 (d, J = 7.4 Hz, 2H, 2CH), 7.51 (d, J = 7.3 Hz, 2H, 2CH), 7.49 (d, J = 7.2 Hz, 2H, 2CH), 7.36 (t, J = 7.2 Hz, 2H, 2CH), 7.21 (t, J = 7.4 Hz, 2H, 2CH), 7.08 (t, J = 7.2 Hz, 1H, CH), 6.89 (t, J = 7.1 Hz, 1H, CH), 4.48 (s, 2H, NH2), 2.48 (s, 3H, CH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.2, 148.7, 140.2, 139.8, 139.7, 138.2 and 135.5 (7C), 131.3 (2CH), 130.2 (2CH), 130.1 (C), 129.4 (2CH), 128.9 (2CH), 128.5 (2CH), 128.3 (2CH), 128.1, 127.8 and 122.9 (3C), 122.5 and 120.2 (2CH), 118.3 (2CH), 116.4 (2CH), 85.2 (C), 20.7 (CH3). Anal. Calcd for C32H25BrN6O (589.49): C, 65.20; H, 4.27; N, 14.26. Found: C, 65.28; H, 4.18; N, 14.38%.
6-Amino-5-(4-methoxyphenyl)-N,7-diphenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3e)
Dark yellow solid; yield: 68%, mp 257–259 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.93 (s, 1H, amide NH), 9.23 (s, 1H, NH), 8.02 (d, J = 7.4 Hz, 2H, 2CH), 7.81 (d, J = 7.0 Hz, 2H, 2CH), 7.70–7.57 (m, 5H, 5CH), 7.56 (d, J = 7.0 Hz, 2H, 2CH), 7.35 (t, J = 7.1 Hz, 2H, 2CH), 7.28–7.16 (m, 4H, 4CH), 7.07 (t, J = 7.1 Hz, 2H, 2CH), 6.89 (t, J = 7.2 Hz, 2H, 2CH), 4.40 (s, 2H, NH2), 3.91 (s, 3H, OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.1, 154.4, 149.7, 140.3, 139.7, 138.2 and 136.3 (7C), 131.2 (2CH), 130.6 (C), 129.9 (CH), 128.5 (2CH), 128.4 (2 × 2CH), 128.0 (2CH), 127.7 (C), 122.5 and 120.1 (2CH), 119.3 (C), 118.2 (2CH), 116.4 (2CH), 113.7 (2CH), 85.1 (C), 54.9 (OCH3). Anal. Calcd for C32H26N6O2 (526.60): C, 72.99; H, 4.98; N, 15.96. Found: C, 73.08; H, 4.82; N, 16.02%.
6-Amino-7-(4-chlorophenyl)-5-(4-methoxyphenyl)-N-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3f)
Dark yellow solid; yield: 73%, mp 237–238 °C. IR (KBr) (νmax/cm–1): 3446–3218 (2NH and NH2), 1658 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.84 (s, 1H, amide NH), 9.25 (s, 1H, NH), 8.04 (d, J = 7.7 Hz, 2H, 2CH), 7.80 (d, J = 7.8 Hz, 2H, 2CH), 7.68 (d, J = 7.4 Hz, 2H, 2CH), 7.62 (d, J = 7.8 Hz, 2H, 2CH), 7.54 (d, J = 7.5 Hz, 2H, 2CH), 7.42–7.15 (m, 6H, 6CH), 7.08 (t, J = 7.1 Hz, 1H, CH), 6.89 (t, J = 7.2 Hz, 1H, CH), 4.44 (s, 2H, NH2), 3.91 (s, 3H, OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.2, 154.9, 148.4, 140.2, 139.7, 138.2, 135.1 and 134.1 (8C) 131.2 (2CH) 130.8 (C), 130.0 (2CH), 128.53 (2CH), 128.49 (2CH), 128.35 (2CH), 127.8 (C), 122.5 and 120.1 (2CH), 119.2 (C), 118.3 (2CH), 116.4 (2CH), 113.7 (2CH), 85.1 (C), 54.9 (OCH3). Anal. Calcd for C32H25ClN6O2 (561.01): C, 68.51; H, 4.49; N, 14.98. Found: C, 68.58; H, 4.62; N, 15.04%.
6-Amino-5-(4-chlorophenyl)-N,7-diphenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3g)
Dark yellow solid; yield: 90%, mp 285–286 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.89 (s, 1H, amide NH), 9.24 (s, 1H, NH), 8.00 (d, J = 7.4 Hz, 2H, 2CH), 7.87 (d, J = 7.3 Hz, 2H, 2CH), 7.73 (d, J = 7.3 Hz, 2H, 2CH), 7.68–7.56 (m, 5H, 5CH), 7.50 (d, J = 7.2 Hz, 2H, 2CH), 7.35 (t, J = 7.6 Hz, 2H, 2CH), 7.22 (t, J = 7.4 Hz, 2H, 2CH), 7.07 (t, J = 7.4 Hz, 1H, CH), 6.89 (t, J = 7.3 Hz, 1H, CH), 4.53 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.1, 150.1, 140.0, 139.7, 138.2, 136.1 and 134.4 (7C), 131.6 (2CH), 129.5 (CH), 129.0 (C), 128.5 (2CH), 128.43 (2CH), 128.41 (2CH), 128.3 (2CH), 128.0 (2CH), 127.6 and 126.5 (2C), 122.5 and 120.2 (2CH), 118.2 (2CH), 116.4 (2CH), 85.3 (C). Anal. Calcd for C31H23ClN6O (561.02): C, 70.11; H, 4.36; N, 15.82. Found: C, 70.05; H, 4.58; N, 15.94%.
6-Amino-5,7-bis(4-chlorophenyl)-N-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3h)
Dark yellow solid; yield: 86%, mp 281–283 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.79 (s, 1H, amide NH), 9.23 (s, 1H, NH), 8.03 (d, J = 7.6 Hz, 2H, 2CH), 7.85 (d, J = 7.4 Hz, 2H, 2CH), 7.75–7.64 (m, 4H, 4CH), 7.60 (d, J = 7.3 Hz, 2H, 2CH), 7.49 (d, J = 7.4 Hz, 2H, 2CH), 7.38 (t, J = 7.3 Hz, 2H, 2CH), 7.21 (d, J = 7.2 Hz, 2H, 2CH), 7.07 (t, J = 7.1 Hz, 1H, 1CH), 6.88 (t, J = 7.0 Hz, H, CH), 4.57 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.6 (C=O), 155.1, 148.9, 140.0, 139.6, 138.1, 135.0, 134.4 and 134.2 (8C), 131.6 (2CH), 131.1 (C), 130.3 (2CH), 128.5 (2CH), 128.44 (2CH), 128.40 (2CH), 128.3 (2CH), 127.7 and 126.4 (2C), 122.5 and 120.2 (2CH), 118.3 (2CH), 116.4 (2CH), 85.3 (C). Anal. Calcd for C31H22Cl2N6O (565.46): C, 65.85; H, 3.92; N, 14.86. Found: C, 65.93; H, 4.02; N, 14.98%.
6-Amino-5-(4-chlorophenyl)-N-phenyl-2-(phenylamino)-7-(thiophen-2-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3i)
Dark yellow solid; yield: 69%, mp 270–271 °C. IR (KBr) (νmax/cm–1): 3453–3216 (2NH and NH2), 1643 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.80 (s, 1H, amide NH), 9.25 (s, 1H, NH), 8.16–8.12 (m, 1H, CH), 7.96 (d, J = 4.0 Hz, 1H, CH), 7.82 (d, J = 7.8 Hz, 2H, 2CH), 7.80 (d, J = 7.6 Hz, 2H, 2CH), 7.75 (d, J = 7.6 Hz, 2H, 2CH), 7.47 (d, J = 7.5 Hz, 2H, 2CH), 7.41 (t, J = 7.1 Hz, 2H, 2CH), 7.32 (t, J = 3.3 Hz, 1H, CH), 7.21 (t, J = 7.0 Hz, 2H, 2CH), 7.11 (t, J = 7.0 Hz, 1H, CH), 6.88 (t, J = 7.0 Hz, 1H, CH), 4.74 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.4, 143.8, 140.4, 139.8, 139.6, 138.3 and 134.5 (7 C), 131.7 (2CH), 131.0, 130.5 and 129.5 (3CH), 128.6 (2CH), 128.5 (2CH+C), 128.3 (2CH), 126.8 and 126.3 (2C), 122.6 and 120.2 (2CH), 118.1 (2CH), 116.5 (2CH), 84.8 (C). Anal. Calcd for C29H21ClN6OS (537.04): C, 64.86; H, 3.94; N, 15.65. Found: C, 64.79; H, 4.00; N, 15.73%.
6-Amino-7-(4-chlorophenyl)-N-(4-methoxyphenyl)-5-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3j)
Dark yellow solid; yield: 81%, mp 217–218 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.69 (s, 1H, amide NH), 9.28 (s, 1H, NH), 8.05 (d, J = 7.5 Hz, 2H, 2CH), 7.82 (d, J = 7.3 Hz, 2H, 2CH), 7.72–7.62 (m, 5H, 5CH), 7.53 (d, J = 7.4 Hz, 2H, 2CH), 7.49 (d, J = 7.1 Hz, 2H, 2CH), 7.19 (d, J = 7.1 Hz, 2H, 2CH), 6.93 (d, J = 7.0 Hz, 2H, 2CH), 6.86 (t, J = 7.3 Hz, 1H, CH), 4.45 (s, 2H, NH2), 3.75 (s, 3H, OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.4 (C=O), 155.1, 154.7, 148.7, 140.1, 139.7, 135.1, 134.1 and 130.5 (8C), 130.0 (2CH), 129.9 (CH), 129.5 (2CH), 128.4 (2×2CH), 128.3 (2CH), 127.7 and 127.5 (2C), 120.1 (CH), 119.8 (2CH), 116.4 (2CH), 113.7 (2CH), 113.2 and 85.2 (2C), 54.7 (OCH3). Anal. Calcd for C32H25ClN6O2 (561.04): C, 68.50; H, 4.49; N, 14.98. Found: C, 68.43; H, 4.53; N, 15.08%.
6-Amino-7-(4-bromophenyl)-N-(4-methoxyphenyl)-5-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3k)
Dark yellow solid; yield: 78%, mp 256–258 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.70 (s, 1H, amide NH), 9.28 (s, 1H, NH), 8.05 (d, J = 7.6 Hz, 2H, 2CH), 7.81 (d, J = 7.4 Hz, 2H, 2CH), 7.78–7.60 (m, 5H, 5CH), 7.55 (d, J = 7.4 Hz, 2H, 2CH), 7.50 (d, J = 7.2 Hz, 2H, 2CH), 7.20 (d, J = 7.1 Hz, 2H, 2CH), 6.94 (d, J = 7.4 Hz, 2H, 2CH), 6.87 (t, J = 7.2 Hz, 1H, CH), 4.48 (s, 2H, NH2), 3.75 (s, 3H, OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.4 (C=O), 155.1, 154.7, 148.8, 140.1, 139.7 and 135.5 (6C), 131.3 (2CH), 130.6 (C), 130.2 (2CH), 129.9 (CH), 129.5 (2CH), 128.4 (2CH), 128.3 (2CH), 127.7, 127.5 and 122.9 (3C), 120.1 (CH), 119.9 (2CH), 116.4 (2CH), 113.7 (2CH), 113.2 and 85.2 (2C), 54.7 (OCH3). Anal. Calcd for C32H25BrN6O2 (605.49): C, 63.48; H, 4.16; N, 13.88. Found: C, 63.36; H, 4.22; N, 14.02%.
6-Amino-7-(4-chlorophenyl)-N-(4-methoxyphenyl)-2-(phenylamino)-5-p-tolylpyrazolo[1,5-a]pyrimidine-3-carboxamide (3l)
Dark yellow solid; yield: 89%, mp 263–265 °C. 3472–3253 (2NH and NH2), 1672 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.69 (s, 1H, amide NH), 9.26 (s, 1H, NH), 8.03 (d, J = 7.7 Hz, 2H, 2CH), 7.72 (d, J = 7.2 Hz, 2H, 2CH), 7.67 (d, J = 7.5 Hz, 2H, 2CH), 7.56–7.47 (m, 6H, 6CH), 7.20 (d, J = 7.4 Hz, 2H, 2CH), 6.93 (d, J = 7.5 Hz, 2H, 2CH), 6.88 (t, J = 7.1 Hz, 1H, CH), 4.42 (s, 2H, NH2), 3.75 (s, 3H, OCH3), 2.47 (s, 3H, CH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.4 (C=O), 155.1, 154.7, 148.6, 140.2, 139.7, 139.6, 135.1, 134.1 and 130.7 (9C), 130.0 (2CH), 129.3 (2CH), 128.9 (2CH), 128.4 (2×2CH), 127.7 and 124.5 (2C), 120.1 (CH), 119.9 (2CH), 116.4 (2CH), 113.7 (2CH), 113.4 and 85.1 (2C), 54.7 (OCH3), 20.7 (CH3). Anal. Calcd for C33H27ClN6O2 (575.07): C, 68.92; H, 9.89; N, 14.61. Found: C, 68.96; H, 9.93; N, 14.72%.
6-Amino-7-(4-bromophenyl)-N-(4-methoxyphenyl)-2-(phenylamino)-5-p-tolylpyrazolo[1,5-a]pyrimidine-3-carboxamide (3m)
Dark yellow solid; yield: 75%, mp 282–283 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.70 (s, 1H, amide NH), 9.29 (s, 1H, NH), 7.97 (d, J = 7.6 Hz, 2H, 2CH), 7.82 (d, J = 7.4 Hz, 2H, 2CH), 7.73 (d, J = 7.6 Hz, 2H, 2CH), 7.57–7.44 (m, 6H, 6CH), 7.21 (d, J = 7.4 Hz, 2H, 2CH), 6.94 (d, J = 7.7 Hz, 2H, 2CH), 6.88 (t, J = 7.1 Hz, 1H, CH), 4.42 (s, 2H, NH2), 3.75 (s, 3H, OCH3), 2.48 (s, 3H, CH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.4 (C=O), 155.1, 154.7, 148.7, 140.2, 139.7,139.6 and 135.5 (7C), 131.3 (2CH), 130.2 (2CH), 130.1 (C), 129.3 (2CH), 128.9 (2CH), 128.3 (2CH), 128.1, 127.6 and 122.9 (3C), 120.1 (CH), 119.9 (2CH), 116.4 (2CH), 113.7 (2CH), 113.2 and 85.1 (2C), 54.7 (OCH3), 20.7 (CH3). Anal. Calcd for C33H27BrN6O2 (619.52): C, 63.98; H, 4.39; N, 13.57. Found: C, 64.05; H, 4.42; N, 13.68%.
6-Amino-N,5-bis(4-methoxyphenyl)-7-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3n)
Dark yellow solid; yield: 65%, mp 248–250 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.74 (s, 1H, amide NH), 9.26 (s, 1H, NH), 7.98 (d, J = 7.6 Hz, 2H, 2CH), 7.80 (d, J = 7.3 Hz, 2H, 2CH), 7.72–7.56 (m, 7H, 7CH), 7.37–7.16 (m, 4H, 4CH), 6.93 (d, J = 7.6 Hz, 2H, 2CH), 6.89 (t, J = 7.2 Hz, 1H, CH), 4.38 (s, 2H, NH2), 3.89 and 3.75 (2s, 6H, 2OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.1, 155.0, 154.7, 149.7, 140.3, 139.7 and 136.3 (7C), 131.2 (2CH), 130.8 (C), 129.4 (CH), 128.3 (2CH), 128.0 (2CH), 127.7 (C), 120.0 (CH), 119.8 (2CH), 119.3 (C), 118.2 (2CH), 116.3 (2CH), 113.68 (2CH), 113.66 (2CH), 113.3 and 85.1 (2C), 54.9 and 54.7 (2OCH3). Anal. Calcd for C33H27N6O3 (555.62): C, 71.34; H, 4.90; N, 15.13. Found: C, 71.42; H, 5.03; N, 15.09%.
6-Amino-7-(4-chlorophenyl)-N,5-bis(4-methoxyphenyl)-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3o)
Dark yellow solid; yield: 79%, mp 268–269 °C. IR (KBr) (νmax/cm–1): 3456–3219 (2NH and NH2), 1663 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.70 (s, 1H, amide NH), 9.27 (s, 1H, NH), 8.04 (d, J = 7.3 Hz, 2H, 2CH), 7.79 (d, J = 7.6 Hz, 2H, 2CH), 7.67 (d, J = 7.5 Hz, 2H, 2CH), 7.57–7.44 (m, 2H, 2CH), 7.50–7.37 (m, 5H, 5CH), 6.93 (d, J = 7.4 Hz, 2H, 2CH), 6.88 (t, J = 7.3 Hz, 1H, CH), 4.48 (s, 2H, NH2), 3.89 and 3.72 (2OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.4 (C=O), 155.14, 155.09, 154.6, 148.5, 140.2, 139.7, 135.2 and 134.0 (8C), 131.2 (2CH), 131.1 (C), 130.0 (2CH), 128.3 (2×2CH), 127.6 (C), 120.1 (CH), 119.8 (2CH), 119.2 (C), 116.4 (2CH), 113.71 (2CH), 113.68 (2CH), 113.1 and 85.6 (2C), 54.8 and 54.7 (2OCH3). Anal. Calcd for C33H27ClN6O3 (591.12): C, 67.05; H, 4.60; N, 14.22. Found: C, 66.96; H, 4.72; N, 14.36%.
6-Amino-5-(4-chlorophenyl)-N-(4-methoxyphenyl)-7-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3p)
Dark yellow solid; yield: 83%, mp 245–246 °C. 3438–3249 (2NH and NH2), 1663 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.74 (s, 1H, amide NH), 9.27 (s, 1H, NH), 8.00 (d, J = 7.8 Hz, 2H, 2CH), 7.87 (d, J = 7.5 Hz, 2H, 2CH), 7.74 (d, J = 7.2 Hz, 2H, 2CH), 7.68–7.58 (m, 3H, 3CH), 7.52 (d, J = 7.3 Hz, 2H, 2CH), 7.50 (d, J = 7.1 Hz, 2H, 2CH), 7.23 (t, J = 7.4 Hz, 2H, 2CH), 6.93 (d, J = 7.1 Hz, 2H, 2CH), 6.89 (t, J = 7.4 Hz, 1H, CH), 4.52 (s, 2H, NH2), 3.75 (s, 3H, OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.4 (C=O), 155.0, 154.7, 150.1, 140.0, 139.7, 136.1 and 134.4 (7C), 131.6 (2CH), 129.5 (CH), 128.9 (C), 128.5 (2CH), 128.41 (2CH), 128.37 (2CH), 128.0 (2CH), 127.9 and 126.5 (2C), 120.1 (CH), 119.9 (2CH), 116.4 (2CH), 113.7 (2CH), 113.2 and 85.2 (2C), 54.7 (OCH3). Anal. Calcd for C32H25ClN6O2 (561.04): C, 68.51; H, 4.49; N, 14.98. Found: C, 68.59; H, 4.56; N, 14.92%.
6-Amino-5,7-bis(4-chlorophenyl)-N-(4-methoxyphenyl)-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3q)
Dark yellow solid; yield: 78%, mp 245–246 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.67 (s, 1H, amide NH), 9.27 (s, 1H, NH), 8.03 (d, J = 7.6 Hz, 2H, 2CH), 7.85 (d, J = 7.4 Hz, 2H, 2CH), 7.73 (d, J = 7.5 Hz, 2H, 2CH), 7.68 (d, J = 7.4 Hz, 2H, 2CH), 7.53 (d, J = 7.5 Hz, 2H, 2CH), 7.50 (d, J = 7.4 Hz, 2H, 2CH), 7.22 (t, J = 7.4 Hz, 2H, 2CH), 6.94 (d, J = 7.6 Hz, 2H, 2CH), 6.89 (t, J = 7.8 Hz, 1H, CH), 4.56 (s, 2H, NH2), 3.76 (s, 3H, OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.4 (C=O), 155.1, 154.7, 148.9, 140.0, 139.7, 135.0, 134.4 and 134.1 (8C), 131.6 (2CH), 131.2 (C), 130.0 (2CH), 128.5 (2CH), 128.4 (2×2CH), 128.0 and 126.5 (2C), 120.2 (CH), 119.9 (2CH), 116.4 (2CH), 113.7 (2CH), 113.2 and 85.2 (2C), 54.7 (OCH3). Anal. Calcd for C32H24Cl2N6O2 (595.53): C, 64.54; H, 4.07; N, 14.11. Found: C, 64.64; H, 3.98; N, 14.03%.
6-Amino-5-(4-chlorophenyl)-N-(4-methoxyphenyl)-2-(phenylamino)-7-(thiophen-2-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3r)
Dark yellow solid; yield: 69%, mp 240–242 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.67 (s, 1H, amide NH), 9.26 (s, 1H, NH), 8.13 (dd, J = 0.6, 3.2 Hz, 1H, CH), 7.92 (d, J = 4.0 Hz, 1H, CH), 7.82 (d, J = 7.7 Hz, 2H, 2CH), 7.74 (d, J = 7.6 Hz, 2H, 2CH), 7.70 (d, J = 7.6 Hz, 2H, 2CH), 7.47 (d, J = 7.5 Hz, 2H, 2CH), 7.34–7.27 (m, 1H, CH), 7.21 (t, J = 7.3 Hz, 2H, 2CH), 6.97 (d, J = 7.5 Hz, 2H, 2CH), 6.88 (t, J = 7.4 Hz, 1H, 1CH), 4.67 (s, 2H, NH2), 3.77 (s, 3H, OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.3 (C=O), 155.3, 154.7, 143.7, 140.5, 139.8, 139.6 and 134.5 (7C), 131.7 (2CH), 131.4 (CH), 131.0 (C), 13 0.4 and 129.4 (2CH), 128.5 (2CH+C), 128.3 (2CH), 126.6 and 126.3 (2C), 120.2 (CH), 119.6 (2CH), 116.4 (2CH), 113.7 (2CH), 113.4 and 84.8 (2C), 54.7 (OCH3). Anal. Calcd for C30H23ClN6O2S (567.11): C, 63.54; H, 4.10; N, 14.82. Found: C, 63.58; H, 4.08; N, 14.88%.
6-Amino-N,7-bis(4-chlorophenyl)-5-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3s)
Dark yellow solid; yield: 92%, mp 274–275 °C. IR (KBr) (νmax/cm–1): 3463–3239 (2NH and NH2), 1658 (C=O). IR (KBr) (νmax/cm–1): 3424–3228 (2NH and NH2), 1652 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.84 (s, 1H, amide NH), 9.17 (s, 1H, NH), 8.05 (d, J = 7.7 Hz, 2H, 2CH), 7.80 (d, J = 7.3 Hz, 2H, 2CH), 7.74–7.56 (m, 7H, 7CH), 7.47 (d, J = 7.1Hz, 2H, 2CH), 7.37 (d, J = 7.9 Hz, 2H, 2CH), 7.18 (t, J = 7.1 Hz, 2H, 2CH), 7.68 (t, J = 7.4 Hz, 1H, CH), 4.47 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.6 (C=O), 155.2, 148.8, 140.1, 139.6, 137.1, 135.0, 134.2 and 130.3 (8C), 130.0 (2CH), 129.9 (CH), 129.4 (2CH), 128.40 (2CH), 128.36 (2CH), 128.35 (2CH), 127.4, 127.3 and 126.1 (3C), 120.2 (CH), 119.85 (2CH), 119.83 (2CH), 116.5 (2CH), 85.1 (C). Anal. Calcd for C31H22Cl2N6O (595.53): C, 65.85; H, 3.92; N, 14.86. Found: C, 65.91; H, 3.86; N, 14.94%.
6-Amino-7-(4-bromophenyl)-N-(4-chlorophenyl)-5-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3t)
Dark yellow solid; yield: 85%, mp 294–296 °C. IR (KBr) (νmax/cm−1): 3453–3219 (2NH and NH2), 1658 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.86 (s, 1H, amide NH), 9.18 (s, 1H, NH), 7.99 (d, J = 7.2 Hz, 2H, 2CH), 7.83 (d, J = 7.5 Hz, 2H, 2CH), 7.81 (d, J = 7.4 Hz, 2H, 2CH), 7.73–7.58 (m, 5H, 5CH), 7.49 (d, J = 7.2 Hz, 2H, 2CH), 7.40 (d, J = 7.4 Hz, 2H, 2CH), 7.19 (t, J = 7.1 Hz, 2H, 2CH), 6.88 (t, J = 7.3 Hz, 1H, CH), 4.48 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.3, 148.9, 140.2, 139.6, 137.1 and 135.4 (6C), 131.4 (2CH), 130.2 (2CH), 129.9 (CH), 129.4 (2CH), 128.4 (2×2CH), 128.3, 127.7, 127.4, 126.1 and 123.0 (5C), 120.3 (CH), 119.93 (2CH), 119.92 (2CH), 116.5 (2CH), 85.5 (C). Anal. Calcd for C31H22BrClN6O (609.95): C, 61.04; H, 3.64; N, 13.78. Found: C, 60.96; H, 3.72; N, 13.86%.
6-Amino-N,7-bis(4-chlorophenyl)-2-(phenylamino)-5-p-tolylpyrazolo[1,5-a]pyrimidine-3-carboxamide (3u)
Dark yellow solid; yield: 85%, mp 281–282 °C. IR (KBr) (νmax/cm−1): 3448–3225 (2NH and NH2), 1672 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.90 (s, 1H, amide NH), 9.20 (s, 1H, NH), 8.07 (d, J = 7.3 Hz, 2H, 2CH), 7.73 (d, J = 7.4 Hz, 2H, 2CH), 7.70 (d, J = 7.7 Hz, 2H, 2CH), 7.66 (d, J = 7.8 Hz, 2H, 2CH), 7.53 (d, J = 7.8 Hz, 2H, 2CH), 7.51 (d, J = 7.3 Hz, 2H, 2CH), 7.42 (d, J = 7.7 Hz, 2H, 2CH), 7.22 (t, J = 7.1 Hz, 2H, 2CH), 6.90 (t, J = 7.2 Hz, 1H, CH), 4.51 (s, 2H, NH2), 2.49 (s, 3H, CH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.6 (C=O), 155.0, 150.1, 140.2, 139.7, 139.6, 137.1, 135.1, 134.1 and 130.8 (9C), 130.0 (2CH), 129.3 (2CH), 128.9 (2CH), 128.43 (2CH), 128.38 (2CH), 127.1, 126.1 and 124.6 (3C), 120.2 (CH), 119.94 (2CH), 119.90 (2CH), 116.5 (2CH), 85.2 (C), 20.8 (CH3). Anal. Calcd for C32H24Cl2N6O (579.53): C, 66.32; H, 4.18; N, 14.50. Found: C, 66.25; H, 4.24; N, 14.64%.
6-Amino-7-(4-bromophenyl)-N-(4-chlorophenyl)-2-(phenylamino)-5-p-tolylpyrazolo[1,5-a]pyrimidine-3-carboxamide (3v)
Dark yellow solid; yield: 82%, mp 271–273 °C. IR (KBr) (νmax/cm−1): 3461–3227 (2NH and NH2), 1671 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.88 (s, 1H, amide NH), 9.20 (s, 1H, NH), 7.99 (d, J = 7.6 Hz, 2H, 2CH), 7.83 (d, J = 7.4 Hz, 2H, 2CH), 7.72 (d, J = 7.3 Hz, 2H, 2CH), 7.66 (d, J = 7.4 Hz, 2H, 2CH), 7.57–7.46 (m, 4H, 4CH), 7.40 (d, J = 7.3 Hz, 2H, 2CH), 7.22 (t, J = 7.0 Hz, 2H, 2CH), 6.90 (t, J = 7.2 Hz, 1H, CH), 4.49 (s, 2H, NH2), 2.48 (s, 3H, CH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.2, 148.6, 140.2, 139.7, 139.6, 137.1 and 135.1 (7C), 131.3 (2CH), 130.3 (2CH), 130.0 (C), 129.3 (2CH), 128.9 (2CH), 128.4 (2CH), 128.0, 127.5, 126.6 and 123.0 (4C), 120.3 (CH), 119.93 (2CH), 119.91 (2CH), 116.5 (2CH), 84.3 (C), 20.7 (CH3). Anal. Calcd for C32H24BrClN6O (609.95): C, 61.60; H, 3.88; N, 13.47. Found: C, 61.68; H, 3.94; N, 13.54%.
6-Amino-N-(4-chlorophenyl)-5-(4-methoxyphenyl)-7-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3w)
Dark yellow solid; yield: 75%, mp 255–257 °C. IR (KBr) (νmax/cm−1): 3468–3226 (2NH and NH2), 1656 (C=O). 1H NMR (500.1 MHz, DMSO-d6): δ 9.96 (s, 1H, amide NH), 9.18 (s, 1H, NH), 8.03 (d, J = 7.5 Hz, 2H, 2CH), 7.82 (d, J = 7.2 Hz, 2H, 2CH), 7.74–7.16 (m, 13H, 13CH), 6.88 (t, J = 7.3 Hz, 1H, CH), 4.42 (s, 2H, NH2), 3.92 (s, 3H, OCH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.7 (C=O), 155.0, 154.8, 150.1, 140.3, 139.7, 137.1 and 136.2 (7C), 131.2 (2CH), 130.8 (C), 129.4 (CH), 128.4 (2CH), 128.3 (2CH), 127.6 and 126.8 (2C), 120.2 (CH), 119.88 (2CH), 119.87 (2CH), 119.2 (C), 118.81 (2CH), 116.6 (2CH), 113.7 (2CH), 85.3 (C), 54.9 (OCH3). Anal. Calcd for C32H25ClN6O2 (561.04): C, 68.51; H, 4.49; N, 14.98. Found: C, 68.46; H, 4.57; N, 15.07%.
6-Amino-N,5-bis(4-chlorophenyl)-7-phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3x)
Dark yellow solid; yield: 89%, mp 255–257 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.89 (s, 1H, amide NH), 9.16 (s, 1H, NH), 8.00 (d, J = 7.5 Hz, 2H, 2CH), 7.85 (d, J = 7.3 Hz, 2H, 2CH), 7.72 (d, J = 7.4 Hz, 2H, 2CH), 7.68–7.55 (m, 5H, 5CH), 7.48 (d, J = 7.2 Hz, 2H, 2CH), 7.36 (d, J = 7.5 Hz, 2H, 2CH), 7.21 (t, J = 7.2 Hz, 2H, 2CH), 6.89 (t, J = 7.0 Hz, 1H, CH), 4.52 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.6 (C=O), 155.0, 150.1, 140.0, 139.6, 137.1, 136.0 and 134.4 (7C), 131.6 (2CH), 129.5 (CH), 129.1 (C), 128.4 (2CH), 128.32 (2CH), 128.25 (2CH), 128.0 (2CH), 127.8, 126.4 and 126.0 (3C), 120.2 (CH), 119.73 (2CH), 119.71 (2CH), 116.4 (2CH), 85.5 (C). Anal. Calcd for C31H22Cl2N6O (595.53): C, 65.85; H, 3.92; N, 14.86. Found: C, 65.79; H, 4.02; N, 14.78%.
6-Amino-N,5,7-tris(4-chlorophenyl)-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3y)
Dark yellow solid; yield: 88%, mp 268–270 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.87 (s, 1H, amide NH), 9.20 (s, 1H, NH), 8.06 (d, J = 7.6 Hz, 2H, 2CH), 7.86 (d, J = 7.5 Hz, 2H, 2CH), 7.78–7.58 (m, 4H, 4CH), 7.52–7.33 (m, 6H, 6CH), 7.26 (t, J = 7.2 Hz, 2H, 2CH), 6.88 (t, J = 7.1 Hz, 2H, 2CH), 4.65 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.4 (C=O), 155.1, 148.9, 140.0, 139.7, 137.3, 135.0, 134.4, 134.1 and 131.6 (9C), 131.2 (2CH), 130.0 (2CH), 128.5 (2CH), 128.4 (2CH), 128.0 (2CH), 127.7, 126.5 and 126.2 (3C), 120.2 (CH), 119.92 (2CH), 119.86 (2CH), 116.4 (2CH), 85.2 (C). Anal. Calcd for C31H21Cl3N6O (599.91): C, 62.07; H, 3.53; N, 14.01. Found: C, 61.98; H, 3.62; N, 14.12%.
6-Amino-N,5-bis(4-chlorophenyl)-2-(phenylamino)-7-(thiophen-2-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide (3z)
Dark yellow solid; yield: 73%, mp 274–276 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 9.78 (s, 1H, amide NH), 9.14 (s, 1H, NH), 8.14–8.05 (m, 1H, CH), 7.95–7.86 (m, 1H, CH), 7.79 (d, J = 7.6 Hz, 2H, 2CH), 7.76 (d, J = 7.6 Hz, 2H, 2CH), 7.72 (d, J = 7.5 Hz, 2H, 2CH), 7.43 (d, J = 7.6 Hz, 2H, 2CH), 7.39 (d, J = 7.5 Hz, 2H, 2CH), 7.33–7.27 (m, 1H, CH), 7.19 (d, J = 7.4 Hz, 2H, 2CH), 6.87 (t, J = 7.2 Hz, 1H, CH), 4.67 (s, 2H, NH2). 13C NMR (125.1 MHz, DMSO-d6): δ 161.5 (C=O), 155.3, 143.7, 140.4, 139.8, 139.5, 137.2 and 134.5 (7C), 131.7 (2CH), 130.9, 130.4 and 129.5 (3CH), 128.5 (2CH+C), 128.3 (2CH), 126.8, 126.2 and 126.0 (3C), 120.2 (CH), 119.60 (2CH), 119.59 (2CH), 116.4 (2CH), 84.7 (C). Anal. Calcd for C29H20Cl2N6OS (571.49): C, 60.95; H, 3.53; N, 14.71. Found: C, 61.05; H, 3.68; N, 14.86%.
General Procedure for the Synthesis of Highly Substituted Ethyl 6-Amino-5,7-Diaryl-2-Phenylpyrazolo[1,5-a]Pyrimidine-3-Carboxylate (3aa-ai)
A mixture of α-azidochalcone 1 (1 mmol), ethyl 3-amino-5-phenyl-1H-pyrazole-4-carboxylate 2d (2 mmol), and triethylamine (Et3N) (2 mmol) in tetrahydrofurane (THF) (4 mL) was stirred at ambient temperature for almost 20 min. After completion of the reaction (confirmed by TLC monitoring), the solvent was evaporated under the low pressure. Then, the residue was recrystallized from EtOAc to obtain desirable products 3aa-ai.
Ethyl 6-amino-7-(4-chlorophenyl)-2,5-diphenylpyrazolo[1,5-a]pyrimidine-3-carboxylate (3aa)
Pale yellow solid, yield: 87%, mp 180–182 °C. IR (KBr) (νmax/cm–1): 3420 and 3376 (NH2), 1723 (C=O). 1H NMR (500.1 MHz, CDCl3): δ 7.94 (d, J = 8.5 Hz, 2H), 7.78–7.71 (m, 4H, 4CH), 7.62 (t, J = 7.5 Hz, 2H, 2CH), 7.56 (t, J = 7.5 Hz, 1H, CH), 7.52 (d, J = 8.5 Hz, 2H, 2CH), 7.41–7.36 (m, 3H, 3CH), 4.34 (q, J = 7.1 Hz, 2H, OCH2), 3.74 (s, 2H, NH2), 1.30 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (125.1 MHz, CDCl3): δ 163.4 (C=O), 157.1, 151.5, 144.9, 136.2, 135.3 and 133.0 (6C), 130.5 (CH+C), 130.4 (2CH), 130.0 (2CH), 129.9 (2CH), 129.4 (2CH), 129.1 (2CH), 128.7 (CH), 127.9 (C), 127.6 (2CH+C), 99.9 (C), 60.0 (OCH2), 14.2 (CH3). Anal. Calcd for C27H21ClN4O2 (468.94): C, 69.16; H, 4.51; N, 11.95. Found: C, 69.23; H, 4.58; N, 12.04%.
Ethyl 6-amino-7-(4-bromophenyl)-2,5-diphenylpyrazolo[1,5-a]pyrimidine-3-carboxylate (3ab)
Pale yellow solid, yield: 76%, mp 201–202 °C. 1H NMR (500.1 MHz, CDCl3): δ 7.88 (d, J = 8.3 Hz, 2H, 2CH), 7.78–7.72 (m, 4H, 4CH), 7.70 (d, J = 8.3 Hz, 2H, 2CH), 7.63 (t, J = 7.6 Hz, 2H, 2CH), 7.57 (t, J = 7.4 Hz, 1H, CH), 7.46–7.37 (m, 3H, 3CH), 4.35 (q, J = 7.1 Hz, 2H, OCH2), 3.72 (s, 2H, NH2), 1.30 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (125.1 MHz, CDCl3): δ 163.4 (C=O), 157.1, 151.6, 145.0, 135.7 and 133.0 (5C), 132.1 (2CH), 130.63 (2CH), 130.55 (CH), 130.00 (2CH), 129.95 (2CH), 129.8 (C), 129.4 (2CH), 128.7 (CH), 127.9 (C), 127.64 (2CH), 127.56, 124.6 and 100.0 (3C), 60.1 (OCH2), 14.2 (CH3). EI-MS, m/z (%): 516 (M+ 81Br, 98), 514 (M+ 79Br, 100), 469 (29), 467 (30), 442 (46), 440 (46), 367 (10), 185 (14), 155 (21), 105 (26), 89 (7), 77 (23). Anal. Calcd for C27H21BrN4O2 (513.39): C, 63.17; H, 4.12; N, 10.91. Found: C, 63.03; H, 4.04; N, 10.82%.
Ethyl 6-amino-7-(4-chlorophenyl)-2-phenyl-5-p-tolylpyrazolo[1,5-a]pyrimidine-3-carboxylate (3ac)
Pale yellow solid, yield: 90%, mp 221–223 °C 1H NMR (500.1 MHz, CDCl3): δ 7.94 (d, J = 8.5 Hz, 2H, 2CH). 7.75–7.73 (m,2H, 2CH), 7.65 (d, J = 8.0 Hz, 2H, 2CH), 7.53 (d, J = 8.5 Hz, 2H, 2CH), 7.43 (d, J = 8.0 Hz, 2H, 2CH), 7.40–7.36 (m, 3H, 3CH), 4.34 (q, J = 7.1 Hz, 2H, OCH2), 3.73 (s, 2H, NH2), 2.47 (s, 3H, CH3), 1.30 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (125.1 MHz, CDCl3): δ 163.4 (C=O), 157.1, 151.4, 145.0, 140.8, 136.2 and 133.1 (6C), 130.4 (2CH), 130.12 (2CH), 130.05 (C), 129.97 (2CH), 129.8 (2CH), 129.1 (2CH), 128.7 (CH), 127.61 (2CH), 127.56, 126.8, 124.8 and 106.5 (4C), 60.1 (OCH2), 21.61 (CH3), 14.2 (CH3). Anal. Calcd for C28H23ClN4O2 (482.97): C, 69.63; H, 4.80; N, 11.60. Found: C, 69.73; H, 4.93; N, 11.65%.
Ethyl 6-amino-7-(4-bromophenyl)-2-phenyl-5-p-tolylpyrazolo[1,5-a]pyrimidine-3-carboxylate (3ad)
Pale yellow solid, yield: 82%, mp 230–232 °C. IR (KBr) (νmax/cm–1): 3458 and 3356 (NH2), 1735 (C=O). 1H NMR (500.1 MHz, CDCl3): δ 7.88 (d, J = 8.5 Hz, 2H, 2CH), 7.79–7.72 (m, 2H, 2CH), 7.70 (d, J =8.5 Hz, 2H, 2CH), 7.65 (d, J = 8.0 Hz, 2H, 2CH), 7.43 (d, J = 8.0 Hz, 2H, 2CH), 7.45–7.33 (m, 3H, 3CH), 4.34 (q, J =7.1 Hz, 2H, OCH2), 3.72 (s, 2H, NH2), 2.48 (s, 3H, CH3), 1.30 (t, J = 7.1 Hz, 3H, CH3).13C NMR (125.1 MHz, CDCl3): δ 163.4 (C=O), 157.1, 151.5, 145.0, 140.8, 135.8 and 133.1 (6C), 132.1 (2CH), 130.7 (2CH), 130.1 (2CH), 130.0 (2CH), 129.9 (C), 129.8 (2CH), 128.7 (CH), 127.6 (2CH), 127.5, 124.9, 124.5 and 99.9 (4C), 60.0 (OCH2), 21.6 (CH3), 14.2 (CH3). Anal. Calcd for C28H23BrN4O2 (527.42): C, 63.76; H, 4.40; N, 10.62. Found: C, 63.84; H, 4.48; N, 10.78%.
Ethyl 6-amino-5-(4-methoxyphenyl)-2,7-diphenylpyrazolo[1,5-a]pyrimidine-3-carboxylate (3ae)
Pale yellow solid, yield: 71%, mp 189–190 °C. IR (KBr) (νmax/cm−1): 3443 and 3362 (NH2), 1736 (C=O). 1H NMR (500.1 MHz, CDCl3): δ 8.31 (d, J = 7.6 Hz, 2H, 2CH), 8.20 (d, J = 7.9 Hz, 2H, 2CH), 7.87 (d, J = 7.9 Hz, 2H, 2CH), 7.68–7.55 (m, 3H, 3CH), 7.50–7.40 (m, 3H, 3CH), 7.11 (d, J = 7.9 Hz, 2H, 2CH), 4.44 (q, J = 7.0 Hz, 2H, CH2), 3.84 (s, 2H, NH2), 3.93 (s, 3H, OCH3), 1.42 (t, J = 7.0 Hz, 3H, CH3). 13C NMR (125.1 MHz, CDCl3): δ 163.5 (C=O), 158.7, 158.4, 150.6, 146.9, 137.1 and 132.9 (6C), 131.4 (2CH), 130.9 (CH), 130.0 (2CH), 129.02 (CH), 128.98 (2CH), 127.8 (2CH), 127.6 (2CH+C), 122.9 and 119.8 (2C), 114.2 (2CH), 105.5 (C), 60.2 (OCH2), 55.5 (OCH3), 14.3 (CH3). Anal. Calcd for C28H24N4O3 (464.52): C, 72.40; H, 5.20; N, 12.06. Found: C, 72.48; H, 5.16; N, 11.98%.
Ethyl 6-amino-7-(4-chlorophenyl)-5-(4-methoxyphenyl)-2-phenylpyrazolo[1,5-a]pyrimidine-3-carboxylate (3af)
Pale yellow solid, yield: 86%, mp 226–228 °C. 1H NMR (500.1 MHz, CDCl3): δ 7.94 (d, J = 8.4 Hz, 2H, 2CH), 7.80–7.72 (m, 4H, 4CH), 7.73 (d, J = 8.7 Hz, 2H, 2CH), 7.53 (d, J = 8.4 Hz, 2H, 2CH), 7.42–7.37 (m, 3H, 3CH), 7.13 (d, J = 8.7 Hz, 2H, 2CH), 4.34 (q, J = 7.1 Hz, 2H, OCH2), 3.92 (s, 3H, OCH3), 3.74 (s, 2H, NH2), 1.30 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (125.1 MHz, CDCl3): δ 161.1 (C=O), 157.1, 151.4, 151.0, 145.0, 136.2, 135.3 and 133.1 (7C), 131.6 (2CH), 130.4 (2CH), 129.96 (2CH), 129.93 (C), 129.1 (2CH), 128.7 (CH), 127.62 (2CH), 127.56 and 119.7 (2C), 114.8 (2CH), 100.5 (C), 60.01 (OCH2), 55.4 (OCH3), 14.2 (CH3). Anal. Calcd for C28H23ClN4O3 (498.97): C, 67.40; H, 4.64; N, 11.23. Found: C, 67.48; H, 4.73; N, 11.32%.
Ethyl 6-amino-5-(4-chlorophenyl)-2,7-diphenylpyrazolo[1,5-a]pyrimidine-3-carboxylate (3ag)
Pale yellow solid, yield: 73%, mp 238–239 °C. 1H NMR (500.1 MHz, DMSO-d6): δ 7.86 (d, J = 7.6 Hz, 2H, 2CH), 7.76 (d, J = 7.9 Hz, 2H, 2CH), 7.68 (d, J = 7.9 Hz, 2H, 2CH), 7.60–7.53 (m, 5H, 5CH), 7.45–7.35 (m, 3H, 3CH), 4.59 (s, 2H, NH2), 4.18 (q, J = 7.1 Hz, 2H, OCH2), 1.75 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (125.1 MHz, DMSO-d6): δ 161.9 (C=O), 154.6, 152.5, 142.8, 136.3, 134.1 and 132.4 (6C), 131.8 (2CH), 129.2 (CH), 128.9 (2CH+C), 128.7 (2CH) 128.2 (2CH), 128.1 (2CH), 128.0 (CH), 127.1 (2CH), 126.7, 126.5 and 98.3 (3C), 58.8 (OCH2), 13.5 (CH3). Anal. Calcd for C27H21ClN4O2 (468.94): C, 69.16; H, 4.51; N, 11.95. Found: C, 69.08; H, 4.43; N, 12.08%.
Ethyl 6-amino-5,7-bis(4-chlorophenyl)-2-phenylpyrazolo[1,5-a]pyrimidine-3-carboxylate (3ah)
Pale yellow solid, yield: 92%, mp 245–246 °C. IR (KBr) (νmax/cm–1): 3449 and 3357 (NH2), 1732 (C=O). 1H NMR (500.1 MHz, CDCl3): δ 7.93 (d, J = 8.5 Hz, 2H, 2CH), 7.79–7.70 (m, 4H, 4CH), 7.61 (d, J = 8.5 Hz, 2H, 2CH), 7.55 (d, J = 8.4 Hz, 2H, 2CH), 7.45–7.37 (m, 3H, 3CH), 4.34 (q, J = 7.1 Hz, 2H, OCH2), 3.74 (s, 2H, NH2),1.30 (t, J = 7.1 Hz, 3H, CH3). 13C NMR (125.1 MHz, CDCl3): δ 163.3 (C=O), 157.1, 151.6, 145.0, 136.7, 136.4, 135.1 and 132.9 (7C), 131.6 (2CH), 130.4 (2CH), 129.9 (2CH), 129.8 (2CH), 129.2 (2CH), 128.8 (CH), 128.3 and 128.2 (2C), 127.7 (2CH), 127.6 and 100.2 (2C), 60.1 (OCH2), 14.2 (CH3). EI-MS, m/z (%): 506 (M+ 37Cl2, 69), 504 (M+ 37Cl35Cl, 69), 502 (M+ 35Cl2, 96), 457 (38), 430 (67), 355 (19), 265 (9), 237 (14), 189 (10), 155 (41), 138 (24), 123 (14), 114 (14), 105 (54), 89 (29), 77 (100), 63 (7), 51 (19). Anal. Calcd for C27H20Cl2N4O2 (503.39): C, 64.42; H, 4.00; N, 11.13. Found: C, 64.39; H, 4.11; N, 11.08%.
Ethyl 6-amino-5-(4-chlorophenyl)-2-phenyl-7-(thiophen-2-yl)pyrazolo[1,5-a]pyrimidine-3-carboxylate (3ai)
Pale yellow solid, yield: 68%, mp 192–193 °C. 1H NMR (500.1 MHz, CDCl3): δ 8.49 (dd, J = 0.9, 3.9 Hz, 1H, CH), 8.25 (d, J = 8.7 Hz, 2H, 2CH), 8.00 (dd, J = 2.2, 7.9 Hz, 2H, 2CH), 7.80 (dd, J = 1.0, 5.0 Hz, 1H, CH), 7.65–7.50 (m, 5H, 5CH), 7.36–7.30 (m, 1H, CH), 4.45 (q, J = 7.0 Hz, 2H, OCH2), 3.78 (s, 2H, NH2), 1.42 (t, J = 7.0 Hz, 3H, CH3). 13C NMR (125.1 MHz, CDCl3): δ 163.4 (C=O), 156.6, 150.3, 140.5, 137.2, 135.5, 133.38, 133.36 and 132.6 (8C), 132.2 (CH), 131.0 (C), 130.0 (2CH), 129.3 (CH), 129.2 (2CH), 128.8 (2CH), 128.7 (CH), 127.93 (2CH), 127.86 (CH), 102.0 (C), 60.4 (OCH2), 14.3 (CH3). Anal. Calcd for C25H19ClN4O2S (474.97): C, 63.22; H, 4.03; N, 11.80. Found: C, 63.31; H, 4.12; N, 11.86%.
α-Glucosidase inhibition assay
α-Glucosidase enzyme (EC3.2.1.20, Saccharomyces cerevisiae, 20 U/mg) and substrate (p-nitrophenyl glucopyranoside) were purchased from Sigma-Aldrich. Enzymewas prepared in potassium phosphate buffer (pH 6.8, 50 mM), and 6-amino-pyrazolo[1,5-a]pyrimidine derivatives 3 were dissolved in DMSO (10% final concentration). The various concentrations of compounds 3 (20 mL), enzyme solution (20 mL) and potassium phosphate buffer (135 mL), were added in the 96-well plate and incubated at 37 °C for 10 min. Then, the substrate (25 mL, 4 mM) was added to the mentioned mixture and allowed to incubate at 37 °C for 20 min. Finally, the change in absorbance was measured at 405 nm by using spectrophotometer (Gen5, Power wave xs2, BioTek, America). DMSO (10% final concentration) and acarbose were used respectively as control and standard drug. The percentage of enzyme inhibitionwas calculated and IC50 values were obtained from non-linear regression curve using the Logit method84.
Kinetic studies
The kinetic analysis were carried out to determine inhibition mode of the most potent compounds (3d and 3af). The 20 mL of enzyme solution (1 U/mL) was incubated with different concentrations (0, 5, 10, and 15 µM) of selected compounds for 15 min at 30 °C. The reaction was then started by adding different concentrations of substrate (p-nitrophenyl glucopyranoside, 1-10 mM), and change in absorbance was measured for 20 min at 405 nm by using spectrophotometer (Gen5, Power wave xs2, BioTek, America).
Molecular docking studies
Autodock 4.2.6 program was used to determine the probable binding conformations of the compounds 3d and 3af over the α-glucosidase active site. AutoDockTools 1.5.2 (ADT) was utilized to prepare the input files. The 3D structure of the most active compounds were drawn 3d and 3af using MarvineSketch 5.8.3, 2012, ChemAxon (http://www.chemaxon.com) and converted to pdbqt coordinate using Auto dockTools85. In AUTOGRID for each atom type in the ligand, maps were calculated with 0.375 Å spacing between grid points, and a grid box of 40 × 40 × 40 Å was created at the center of 12.5825, −7.8955, 12.519 in each dimension to determine the ligand- enzyme interactions. Rigid ligand dockings were accomplished for the selected compounds. Of the three different search algorithms suggested by AutoDock 4.2.6, the Lamarckian genetic algorithm (LGA) consisting of 50 runs, 25 × 106 energy evaluations, and 27,000 generations was carried out86. Other docking parameters were set to default. The best interaction of the selected compounds were considered for analyzing and the results were illustrated using Discovery Studio 4.5 Client.
References
Deshpande, A. D., Harris-Hayes, M. & Schootman, M. Epidemiology of Diabetes and Diabetes-Related Complications. Phys. Ther. 88, 1254–1264, https://doi.org/10.2522/ptj.20080020 (2008).
de Boer, I. H. Kidney Disease and Related Findings in the Diabetes Control andComplications Trial/Epidemiology of Diabetes Interventions and Complications Study. Diabetes Care 37, 24–30, https://doi.org/10.2337/dc13-2113 (2014).
Martin, C. L., Albers, J. W. & Pop-Busui, R. Neuropathy and related findings in the diabetes control and complications trial/epidemiology of diabetes interventions and complications study. Diabetes Care 37, 31–38, https://doi.org/10.2337/dc13-2114 (2014).
Vinholes, J. & Vizzotto, M. Synergisms in alpha-glucosidase inhibition and antioxidant activity of camellia sinensis l. kuntze and eugenia uniflora l. ethanolic extracts. Pharmacognosy Res. 9, 101–107, https://doi.org/10.4103/0974-8490.197797 (2017).
Cho, N. H. et al. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 138, 271–281, https://doi.org/10.1016/j.diabres.2018.02.023 (2018).
Kehm, R. et al. Endogenous advanced glycation end products in pancreatic islets after short-term carbohydrate intervention in obese, diabetes-prone mice. Nutr. Diabetes 9, 9–13, https://doi.org/10.1038/s41387-019-0077-x (2019).
Johnston, P. S. et al. Advantages of α-glucosidase inhibition as monotherapy in elderlytype 2 diabetic patients. J. Clin. Endocrinol. Metab. 83, 1515–1522, https://doi.org/10.1210/jc.83.5.1515 (1998).
David, S. H. & Bell, M. B. Type 2 diabetes mellitus: What is the optimal treatment regimen? Am. J. Med. 116, 23–29, https://doi.org/10.1016/j.amjmed.2003.10.017 (2004).
van de Laar, F. A. Alpha-glucosidase inhibitors in the early treatment of type 2 diabetesn Vasc. Health Risk Manag. 4, 1189–1195, https://doi.org/10.2147/vhrm.s3119 (2008).
Poovitha, S. & Parani, M. In vitro and in vivo α-amylase and α-glucosidase inhibiting activities of the protein extracts from two varieties of bitter gourd (Momordica charantia L.). BMC Complement. Altern. Med. 16, 1–8, https://doi.org/10.1186/s12906-016-1085-1 (2016).
Jacob, G. S. Glycosylation inhibitors in biology and medicine. Curr. Opin. Struct. Biol. 5, 605–611, https://doi.org/10.1016/0959-440x(95)80051-4 (1995).
Dennis, J. W., Laferté, S., Waghorne, C., Breitman, M. L. & Kerbel, R. S. β1-6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science 236, 582–585, https://doi.org/10.1126/science.2953071 (1987).
Asano, N. Glycosidase inhibitors: Update and perspectives on practical use. Glycobiology. 13, 93R–104R, https://doi.org/10.1093/glycob/cwg090 (2003).
Simsek, E. et al. α-Glucosidase inhibitors have a prolonged antiviral effect against hepatitis B virus through the sustained inhibition of the large and middle envelope glycoproteins. Antivir. Chem. Chemother. 17, 259–267, https://doi.org/10.1177/095632020601700503 (2006).
Doseung, L. et al. Antiviral Activity of Methylelaiophylin, an α-Glucosidase Inhibitor. J. Microbiol. Biotechnol. 21, 263–266, https://doi.org/10.4014/jmb.1011.11002 (2011).
Yar, M. et al. Novel synthesis of dihydropyrimidines for α-glucosidase inhibition to treat type 2 diabetes: In vitro biological evaluation and in silico docking. Bioorg. Chem. 54, 96–104, https://doi.org/10.1016/j.bioorg.2014.05.003 (2014).
Zeng, L., Zhang, G., Lin, S. & Gong, D. Inhibitory Mechanism of Apigenin on α-Glucosidase and Synergy Analysis of Flavonoids. J. Agric. Food Chem. 64, 6939–6949, https://doi.org/10.1021/acs.jafc.6b02314 (2016).
Jang, J. H., Park, J. E. & Han, J. S. Scopoletin inhibits α-glucosidase in vitro and alleviates postprandial hyperglycemia in mice with diabetes. Eur. J. Pharmacol. 834, 152–156, https://doi.org/10.1016/j.ejphar.2018.07.032 (2018).
Ding, H. et al. New Insights into the Inhibition Mechanism of Betulinic Acid on α-Glucosidase. J. Agric. Food Chem. 66, 7065–7075, https://doi.org/10.1021/acs.jafc.8b02992 (2018).
Javid, M. T. et al. Synthesis, in vitro α-glucosidase inhibitory potential and molecular docking study of thiadiazole analogs. Bioorg. Chem. 78, 201–209, https://doi.org/10.1016/j.bioorg.2018.03.022 (2018).
Adib, M. et al. New 6-amino-pyrido[2,3-d]pyrimidine-2,4-diones as novel agents to treat type 2 diabetes: A simple and efficient synthesis, α-glucosidase inhibition, molecular modeling and kinetic study. Eur. J. Med. Chem. 155, 353–363, https://doi.org/10.1016/j.ejmech.2018.05.046 (2018).
Adib, M. et al. Design, synthesis and in vitro α-glucosidase inhibition of novel coumarin-pyridines as potent antidiabetic agents. New J. Chem. 42, 17268–17278, https://doi.org/10.1039/c8nj02495b (2018).
Gollapalli, M. et al. Synthesis of benzothiazole derivatives as a potent α-glucosidase inhibitor. Bioorg. Chem. 85, 33–48, https://doi.org/10.1016/j.bioorg.2018.12.021 (2019).
Dhameja, M. & Gupta, P. Synthetic heterocyclic candidates as promising α-glucosidase inhibitors: An overview. Eur. J. Med. Chem. 176, 343–377, https://doi.org/10.1016/j.ejmech.2019.04.025 (2019).
Adib, M. et al. Design and synthesis of new fused carbazole-imidazole derivatives as anti-diabetic agents: In vitro α-glucosidase inhibition, kinetic, and in silico studies. Bioorg. Med. Chem. Lett. 29, 713–718, https://doi.org/10.1016/j.bmcl.2019.01.012 (2019).
Faria, J. V. et al. Recently reported biological activities of pyrazole compounds. Bioorg. Med. Chem. 25, 5891–5903, https://doi.org/10.1016/j.bmc.2017.09.035 (2017).
Nitulescu, G. M., Draghici, C. & Missir, A. V. Synthesis of new pyrazole derivatives and their anticancer evaluation. Eur. J. Med. Chem. 45, 4914–4919, https://doi.org/10.1016/j.ejmech.2010.07.064 (2010).
Koca, I., Özgür, A., Coşkun, K. A. & Tutar, Y. Synthesis and anticancer activity of acyl thioureas bearing pyrazole moiety. Bioorg. Med. Chem. 21, 3859–3865, https://doi.org/10.1016/j.bmc.2013.04.021 (2013).
Malvar, D. D. C. et al. Antinociceptive, anti-inflammatory and antipyretic effects of 1.5-diphenyl-1H-Pyrazole-3-carbohydrazide, a new heterocyclic pyrazole derivative. Life Sci. 95, 81–88, https://doi.org/10.1016/j.lfs.2013.12.005 (2014).
Aly, A. A. Synthesis of Polyfunctionally Substituted Pyrazolonaphthyridine, Pentaazanaphthalene, and Heptaazaphenanthrene Derivatives. Phosphorus, Sulfur, and Silicon 181, 2395–2409, https://doi.org/10.1080/10426500600695179 (2006).
Palazuelos, J. et al. The CB 2 Cannabinoid Receptor Controls Myeloid Progenitor Trafficking. J. Biol. Chem. 283, 13320–13329, https://doi.org/10.1074/jbc.m707960200 (2008).
Newman, A. H. et al. Molecular Determinants of Selectivity and Efficacy at the Dopamine D3 Receptor. J. Med. Chem. 55, 6689–6699, https://doi.org/10.1021/jm300482h (2012).
Chaudhry, F. et al. Evaluation of α-glucosidase inhibiting potentials with docking calculations of synthesized arylidene-pyrazolones. Bioorg. Chem. 77, 507–514, https://doi.org/10.1016/j.bioorg.2018.02.002 (2018).
Ren, L. et al. Potent and selective pyrazolo[1,5-a]pyrimidine based inhibitors of B-Raf V600E kinase with favorable physicochemical and pharmacokinetic properties. Bioorg. Med. Chem. Lett. 22, 1165–1168, https://doi.org/10.1016/j.bmcl.2011.11.092 (2012).
El Sayed, M. T. et al. Tyrosine kinase inhibition effects of novel Pyrazolo[1,5-a]pyrimidines and Pyrido[2,3-d]pyrimidines ligand: Synthesis, biological screening and molecular modeling studies. Bioorg. Chem. 78, 312–323, https://doi.org/10.1016/j.bioorg.2018.03.009 (2018).
Jiang, J. K. et al. Discovery of 3-(4-sulfamoylnaphthyl)pyrazolo[1,5-a]pyrimidines as potent and selective ALK2 inhibitors. Bioorg. Med. Chem. Lett. 28, 3356–3362, https://doi.org/10.1016/j.bmcl.2018.09.006 (2018).
Ali, G. M. E., Ibrahim, D. A., Elmetwali, A. M. & Ismail, N. S. M. Design, synthesis and biological evaluation of certain CDK2 inhibitors based on pyrazole and pyrazolo[1,5-a] pyrimidine scaffold with apoptotic activity. Bioorg. Chem. 86, 1–14, https://doi.org/10.1016/j.bioorg.2019.01.008 (2019).
Almansa, C. et al. Synthesis and SAR of a new series of COX-2-selective inhibitors: Pyrazolo[1,5-α]pyrimidines. J. Med. Chem. 44, 350–361, https://doi.org/10.1021/jm0009383 (2001).
Hwang, J. Y. et al. Discovery and characterization of a novel 7-aminopyrazolo[1,5-a]pyrimidine analog as a potent hepatitis C virus inhibitor. Bioorg. Med. Chem. Lett. 22, 7297–7301, https://doi.org/10.1016/j.bmcl.2012.10.123 (2012).
Sun, L., Gao, P., Zhan, P. & Liu, X. Pyrazolo[1,5-a]pyrimidine-based macrocycles as novel HIV-1 inhibitors: a patent evaluation of WO2015123182. Expert Opin. Ther. Pat. 26, 979–986, https://doi.org/10.1080/13543776.2016.1210127 (2016).
Hassan, A. S., Masoud, D. M., Sroor, F. M. & Askar, A. A. Synthesis and biological evaluation of pyrazolo[1,5-a]pyrimidine-3-carboxamide as antimicrobial agents. Med. Chem. Res. 26, 2909–2919, https://doi.org/10.1007/s00044-017-1990-y (2017).
Abdallah, A. E. M. & Elgemeie, G. H. Design, synthesis, docking, and antimicrobial evaluation of some novel pyrazolo[1,5-a] pyrimidines and their corresponding cycloalkane ring-fused derivatives as purine analogs. Drug Des. Devel. Ther. 12, 1785–1798, https://doi.org/10.2147/DDDT.S159310 (2018).
Fouda, A. M. et al. Synthesis, in vitro antimicrobial and cytotoxic activities of some new pyrazolo[1,5-a]pyrimidine derivatives. Molecules 24, 1080–1099, https://doi.org/10.3390/molecules24061080 (2019).
Tellew, J. E. et al. Discovery of NBI-77860/GSK561679, a potent corticotropin-releasing factor (CRF1) receptor antagonist with improved pharmacokinetic properties. Bioorg. Med. Chem. Lett. 20, 7259–7264, https://doi.org/10.1016/j.bmcl.2010.10.095 (2010).
Childress, E. S. et al. Discovery of Novel Central Nervous System Penetrant Metabotropic Glutamate Receptor Subtype 2 (mGlu 2) Negative Allosteric Modulators (NAMs) Based on Functionalized Pyrazolo[1,5-a]pyrimidine-5-carboxamide and Thieno[3,2-b]pyridine-5-carboxamide Cores. J. Med. Chem. 62, 378–384, https://doi.org/10.1021/acs.jmedchem.8b01266 (2019).
Xu, J. et al. Synthesis and biological evaluation of 7-(2-Chlorophenylamino)-5-((2-[18F]fluoro-ethyoxy)methyl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile as PET tumor imaging agent. Zeitschrift fur Naturforsch. - Sect. B J. Chem. Sci. 67, 827–834, https://doi.org/10.5560/ZNB.2012-0047 (2012).
Metwally, N. H., Mohamed, M. S. & Ragb, E. A. Design, synthesis, anticancer evaluation, molecular docking and cell cycle analysis of 3-methyl-4,7-dihydropyrazolo[1,5-a]pyrimidine derivatives as potent histone lysine demethylases (KDM) inhibitors and apoptosis inducers. Bioorg. Chem. 88, 102929, https://doi.org/10.1016/j.bioorg.2019.102929 (2019).
Balestri, F. et al. Acid Derivatives of Pyrazolo[1,5-a]pyrimidine as Aldose Reductase Differential Inhibitors. Cell Chem. Biol. 25, 1414–1418, https://doi.org/10.1016/j.chembiol.2018.07.008 (2018).
Griffith, D. A. et al. Discovery and evaluation of pyrazolo[1,5-a]pyrimidines as neuropeptide Y1 receptor antagonists. Bioorg. Med. Chem. Lett. 21, 2641–2645, https://doi.org/10.1016/j.bmcl.2010.12.116 (2011).
Ivachtchenko, A. V. et al. Synthesis and structure-activity relationship (SAR) of (5,7-disubstituted 3-phenylsulfonyl-pyrazolo[1,5-a]pyrimidin-2-yl)-methylamines as potent serotonin 5-HT 6 receptor (5-HT 6R) antagonists. J. Med. Chem. 54, 8161–8173, https://doi.org/10.1021/jm201079g (2011)
Ammar, Y. A., Aly, M. M., Al-Sehemi, A. A. G., Salem, M. A. & El-Gaby, M. S. A. Cyanoacetanilides Intermediates in Heterocyclic Synthesis. Part 5: Preparation of Hitherto Unknown 5-Aminopyrazole and Pyrazolo[1,5-a]pyrimidine Derivatives Containing Sulfamoyl Moiety. J. Chinese Chem. Soc. 56, 1064–1071, https://doi.org/10.1002/jccs.200900154 (2009).
Drev, M. et al. Regioselective synthesis of 1- and 4-substituted 7-oxopyrazolo[1,5-a]pyrimidine-3-carboxamides. Tetrahedron 70, 8267–8279, https://doi.org/10.1016/j.tet.2014.09.020 (2014).
Hassan, A. S., Mady, M. F., Awad, H. M. & Hafez, T. S. Synthesis and antitumor activity of some new pyrazolo[1,5-a]pyrimidines. Chinese Chem. Lett. 28, 388–393, https://doi.org/10.1016/j.cclet.2016.10.022 (2017).
Lunagariya, M. V., Thakor, K. P., Waghela, B. N., Pathak, C. & Patel, M. N. Design, synthesis, pharmacological evaluation and DNA interaction studies of binuclear Pt(II) complexes with pyrazolo[1,5-a]pyrimidine scaffold. Appl. Organomet. Chem. 32, 1–25, https://doi.org/10.1002/aoc.4222 (2018).
Castillo, J. C., Tigreros, A. & Portilla, J. 3-Formylpyrazolo[1,5- a]pyrimidines as Key Intermediates for the Preparation of Functional Fluorophores. J. Org. Chem. 83, 10887–10897, https://doi.org/10.1021/acs.joc.8b01571 (2018).
Farag, A. M. & Fahim, A. M. Synthesis, biological evaluation and DFT calculation of novel pyrazole and pyrimidine derivatives. J. Mol. Struct. 1179, 304–314, https://doi.org/10.1016/j.molstruc.2018.11.008 (2019).
Loubidi, M. et al. One-Pot SNAr/Direct Pd-Catalyzed CH Arylation Functionalization of Pyrazolo[1,5-a]pyrimidine at the C3 and C7 Positions. Eur. J. Org. Chem. 2018, 3936–3942, https://doi.org/10.1002/ejoc.201800580 (2018).
Salem, M. A. et al. Recent synthetic methodologies for pyrazolo[1,5-a]pyrimidine. Synth. Commun. 49, 1750–1776, https://doi.org/10.1080/00397911.2019.1604967 (2019).
Modi, P., Patel, S. & Chhabria, M. T. Identification of some novel pyrazolo[1,5-a]pyrimidine derivatives as InhA inhibitors through pharmacophore-based virtual screening and molecular docking. J. Biomol. Struct. Dyn. 37, 1736–1749, https://doi.org/10.1080/07391102.2018.1465852 (2019).
Zhang, X., Song, Y., Gao, L., Guo, X. & Fan, X. Highly facile and regio-selective synthesis of pyrazolo[1,5-a]pyrimidines via reactions of 1,2-allenic ketones with aminopyrazoles. Org. Biomol. Chem. 12, 2099–2107, https://doi.org/10.1039/c3ob42445f (2014).
Shekarrao, K. et al. Microwave-assisted palladium mediated efficient synthesis of pyrazolo[3,4-b]pyridines, pyrazolo[3,4-b]quinolines, pyrazolo[1,5-a]pyrimidines and pyrazolo[1,5-a]quinazolines. RSC Adv. 4, 24001–24006, https://doi.org/10.1039/c4ra02865a (2014).
Jismy, B., Guillaumet, G., Allouchi, H., Akssira, M. & Abarbri, M. Concise and Efficient Access to 5,7-Disubstituted Pyrazolo[1,5-a]pyrimidines by Pd-Catalyzed Sequential Arylation, Alkynylation and SNAr Reaction. Eur. J. Org. Chem. 2017, 6168–6178, https://doi.org/10.1002/ejoc.201701024 (2017).
Jismy, B., Allouchi, H., Guillaumet, G., Akssira, M. & Abarbri, M. An Efficient Synthesis of New 7-Trifluoromethyl-2,5-disubstituted Pyrazolo[1,5-a]pyrimidines. Synth. 50, 1675–1686, https://doi.org/10.1055/s-0036-1591752 (2018).
Chen, W., Hu, M., Wu, J., Zou, H. & Yu, Y. Domino approach for the synthesis of pyrrolo[1,2-α]pyrazine from vinyl azides. Org. Lett. 12, 3863–3865, https://doi.org/10.1021/ol101538x (2010).
Bonnamour, J. & Bolm, C. Iron (II) Triflate as a Catalyst for the Synthesis of Indoles by Intramolecular C-H Amination. Org. Lett. 13, 2012–2014, https://doi.org/10.1021/ol2004066 (2011).
Hu, B. et al. Catalyst-Free Preparation of 1,2,4,5- Tetrasubstituted Imidazoles from a Novel Unexpected Domino Reaction of 2-Azido Acrylates and Nitrones. Org. Lett. 13, 6362–6365, https://doi.org/10.1021/ol202650z (2011).
Shao, J., Yu, W., Shao, Z. & Yu, Y. A “one-pot” multicomponent approach to polysubstituted 4-aminopyridines. Chem. Commun. 48, 2785–2787, https://doi.org/10.1039/c2cc17850h (2012).
Zhang, G. et al. One-pot three-component approach to the synthesis of polyfunctional pyrazoles. Org. Lett. 15, 5967–5969, https://doi.org/10.1021/ol402810f (2013).
Shao, J. et al. Tuning the Annulation Reactivity of Vinyl Azides and Carbazates: A Divergent Synthesis of Aza-pyrimidinones and Imidazoles. Org. Lett. 17, 4502–4505, https://doi.org/10.1021/acs.orglett.5b02180 (2015).
Zhang, G., Chen, B., Guo, X., Guo, S. & Yu, Y. Iron(II)-promoted synthesis of 2-aminothiazoles via C-N bond formation from vinyl azides and potassium thiocyanate. Adv. Synth. Catal. 357, 1065–1069, https://doi.org/10.1002/adsc.201400856 (2015).
Adiyala, P. R., Mani, G. S., Nanubolu, J. B., Shekar, K. C. & Maurya, R. A. Access to Imidazo[1,2-a]pyridines via Annulation of α-Keto Vinyl Azides and 2-Aminopyridines. Org. Lett. 17, 4308–4311, https://doi.org/10.1021/acs.orglett.5b02124 (2015).
Shu, K. et al. Base-mediated synthesis of highly functionalized 2-aminonicotinonitriles from α-keto vinyl azides and α,α-dicyanoalkenes. RSC Adv. 6, 49123–49126, https://doi.org/10.1039/c6ra04669j (2016).
Adib, M., Peytam, F., Rahmanian-Jazi, M., Bijanzadeh, H. R. & Amanlou, M. A newsynthetic strategy towards 2,4,5-trisubstituted 1H-imidazoles and highly substitutedpyrrolo[1,2-c]imidazoles by use of α-azidochalcones via Michael addition-cyclizationfollowed by Wittig reaction. Tetrahedron 73, 6696–6705, https://doi.org/10.1016/j.tet.2017.09.042 (2017).
Adib, M. & Peytam, F. An efficient synthesis of fully substituted pyrazolo[3,4-b]pyridin-5-amines from α-azidochalcones. Tetrahedron 74, 2414–2420, https://doi.org/10.1016/j.tet.2018.03.036 (2018).
Borra, S., Chandrasekhar, D., Newar, U. D. & Maurya, R. A. Access to 2,3-Fused Pyrroles via Visible Light Driven Coupling of α-Azidochalcones with 1/2-Naphthols, or 2-Hydroxy-1,4-Naphthoquinone. J. Org. Chem. 84, 1042–1052, https://doi.org/10.1021/acs.joc.8b02459 (2019).
Daina, A., Michielin, O. & Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 1–13, https://doi.org/10.1038/srep42717 (2017).
Lipinski, C. A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 1, 337–341, https://doi.org/10.1016/j.ddtec.2004.11.007 (2004).
Hughes, J. D. et al. Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorg. Med. Chem. Lett. 18, 4872–4875, https://doi.org/10.1016/j.bmcl.2008.07.071 (2008).
Veber, D. F. et al. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 45, 2615–2623, https://doi.org/10.1021/jm020017n (2002).
Mohammadi-Khanaposhtani, M. et al. New Biscoumarin Derivatives as Potent α-Glucosidase Inhibitors: Synthesis, Biological Evaluation, Kinetic. Analysis, and Docking Study, Polycycl. Aromat. Compd. 38, 1–12, https://doi.org/10.1080/10406638.2018.1509359 (2018).
Nair, V. & George, T. G. A novel synthesis of α-azidocinnamates, α-azido-α,β-unsaturated ketones and β-azidostyrenes mediated by cerium(IV) ammonium nitrate. Tetrahedron Lett. 41, 3199–3201, https://doi.org/10.1016/S0040-4039(00)00350-6 (2000).
Hassan, A. S., Hafez, T. S. & Osman, S. A. Synthesis, characterization, and cytotoxicity of some new 5-aminopyrazole and pyrazolo[1,5-a]pyrimidine derivatives. Sci. Pharm. 83, 27–39, https://doi.org/10.3797/scipharm.1409-14 (2015).
Ghozlan, S. A. S., Abdelrazek, F. M., Mohamed, M. H. & Azmy, K. E. Synthesis of Some New Pyrazole and Pyrazolopyrimidine Derivatives. J. Heterocycl. Chem. 47, 1379–1385, https://doi.org/10.1002/jhet.482 (2010).
Nikookar, H. et al. Design, synthesis and in vitro α-glucosidase inhibition of novel dihydropyrano[3,2-c]quinoline derivatives as potential anti-diabetic agents. Bioorg. Chem. 77, 280–286, https://doi.org/10.1016/j.bioorg.2018.01.025 (2018).
Morris, G. M. et al. Automated docking using a Lamarckian genetic algorithm and anempirical binding free energy function. J. Comput. Chem. 19, 1639–1662, https://doi.org/10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B (1998).
González, J., Giménez, X., Bofill, J. M. Algorithm to Evaluate Rate Constants forPolyatomic Chemical Reactions. II. Applications. J. Comput. Chem. 31, 2111–2121, https://doi.org/10.1002/jcc.20729 (2007).
Acknowledgements
This work was supported and funded by Tehran University of Medical Sciences (TUMS); Gran No. 98-3-263-45230.
Author information
Authors and Affiliations
Contributions
Dr. Alireza Foroumadi and Dr. Mehdi Adib designed the study and conducted the experiments. Fariba Peytam and Reihaneh Shourgeshty synthesized the desired compounds. Fariba Peytam and Mahmoud Rahmanian-Jazi wrote the manuscript, analyzed the characterization data, and prepared the Supporting Information file. Kouros Divsalar, Mohammad Ali Faramarzi, and Somaye Mojtabavi performed the in vitro analysis and kinetic study against α-glucosidase. Loghman Firoozpour carried out the docking studies. Fatemeh Safari evaluated the cytotoxic activity of some selected compounds. Mehdi Jahani, Setareh Moghimi, and Mohammad Mahdavi revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplememtary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peytam, F., Adib, M., Shourgeshty, R. et al. An efficient and targeted synthetic approach towards new highly substituted 6-amino-pyrazolo[1,5-a]pyrimidines with α-glucosidase inhibitory activity. Sci Rep 10, 2595 (2020). https://doi.org/10.1038/s41598-020-59079-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-59079-z
This article is cited by
-
Synthesis, α-Glucosidase inhibitory activity and docking studies of Novel Ethyl 1,2,3-triazol-4-ylmethylthio-5,6-diphenylpyridazine-4-carboxylate derivatives
BMC Chemistry (2023)
-
Imidazo[1,2-c]quinazolines as a novel and potent scaffold of α-glucosidase inhibitors: design, synthesis, biological evaluations, and in silico studies
Scientific Reports (2023)
-
Design, synthesis and α-glucosidase inhibition study of novel pyridazin-based derivatives
Medicinal Chemistry Research (2023)
-
Fused azoloazines with antidiabetic activity
Russian Chemical Bulletin (2022)
-
Design, synthesis, molecular docking, and in vitro α-glucosidase inhibitory activities of novel 3-amino-2,4-diarylbenzo[4,5]imidazo[1,2-a]pyrimidines against yeast and rat α-glucosidase
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.