Abstract
Root-knot nematodes (RKN) such as Meloidogyne spp. are among the most detrimental pests in agriculture affecting several crops. New methodologies to manage RKN are needed such as efficient discovery of nematophagous microbes. In this study, we developed an in vitro high-throughput method relying on the free-living nematode Caenorhabditis elegans and the infection of those nematodes with a soil slurry containing a microbiome likely to house nematophagous microbes. Nematodes were monitored for presence of infection and sub-cultured repeatedly for the purpose of isolating pure cultures of the microbe responsible for conferring the nematicidal activity. Once soil microbes were confirmed to be antagonistic to C. elegans, they were tested for pathogenicity against Meloidogyne chitwoodi. Using this methodology, the fungal isolate Mortierella globalpina was confirmed to be pathogenic in vitro against M. chitwoodi by nematode trapping via hyphal adhesion to the cuticle layer, penetration of the cuticle layer, and subsequently digestion of its cellular contents. M. globalpina was also observed to reduce disease symptomology of RKNs in vivo via significant reduction of root-galls on tomato (Solanum lycopersicum var. Rutgers).
Similar content being viewed by others
Introduction
Members of the phylum Nematoda have been in existence for close to one billion years and are classified as one of the most diverse organisms globally1. Soil-borne nematodes feed on nearly all forms of life, including bacteria, fungi, unicellular eukaryotes, plants, and animals2. Most herbivorous or plant-parasitic nematodes (PPNs) affect crops through feeding within or on plant roots3. However, PPNs of economic importance can be grouped by either causing direct damage to their host or acting as a vector for viruses and/or bacteria3. According to Kaplan, nematodes are annually responsible for ~$100 billion in crop damage and cause 5 percent of all yearly crop loss globally. Meloidogyne spp., or the root-knot nematodes (RKN), are able to infect a wide range of crop families such as Solanaceae, Brassicaceae, Leguminosae, Musaceae, Curcubitaceae, Poaceae, and others4,5. Within their several plant hosts, RKNs possess the ability to dedifferentiate plant cells into multinucleate feeding sites via hypertrophy by synthesizing specific effectors within their esophageal gland. These effectors are then injected into the host plant cell – altering the cell’s DNA to establish a swollen feeding cell5.
Attempts to control nematode pests have been practiced by means of soil sterilization (via fumigation or solarization), crop rotation, and resistance breeding6,7. These control practices have become increasingly difficult over the years. For fumigation, the U.S. Environmental Protection Agency phased out the use of the chemical nematicide methyl bromide in 20058. Introgression of nematode resistance genes is common in rootstock production for many woody plants but can be an impractical or limited process with annual crops due to the few available RKN-resistance genes, and lack of ability to deploy these into the many susceptible crops4,9,10. Crop rotations have served growers little success as RKN members have a wide variety of plant hosts4,11. In organic systems, soil amendments along with solarization practices have been attempted to control RKN, yielding the most success when used simultaneously12. Unfortunately, within these studies - nematode populations near the margins of the solarized beds were observed to recover during the growing season12.
In efforts to combat these difficulties, a limited number of biological products have been formulated that tend to contain Pasteuria nishizawae or Purpureocillium lilacinum strain 25113,14. However, P. nishizawae has only been observed to complete its life cycle in the adults of the soybean cyst nematode. Although P. lilacinum strain 251 is confirmed to have no human risks, other species of P. lilacinum are known to produce mycotoxins and paecilotoxins which are harmful to humans15,16. It should be noted that the reproducibility of nematode control sought after by these commercial microorganisms in agronomic systems appears to be lacking17. Thus, there is a need to isolate new and more efficient microbes that are antagonistic against RKNs.
Plant-parasitic nematodes require a plant host to complete their reproductive cycle, and as such, the establishment of in vitro RKN cultures is a lengthy multi-step process18. The method of culturing RKNs in vitro makes very difficult the development of a high-throughput system to screen for new microbes that could exhibit root knot nematicidal properties. Unlike common PPNs, Caenorhabditis elegans is a model bacterivorous nematode easily grown in vitro19. A series of methodologies have been designed to utilize C. elegans as a model for screening virulence genes of certain human pathogens such as Pseudomonas aeruginosa and several others20,21,22,23. In the present study, it was hypothesized that nematophagous microorganisms would not discriminate against different species of nematodes that practice different feeding habits as the cuticle layers of both bacterivores (Caenorhabditis spp.) and RKNs (Meloidogyne spp.) are largely composed of collagen24. In support of our hypothesis, chemical nematicides such fluensulfone have been found to induce negative effects on both Caenorhabditis and RKNs25. To test this hypothesis, we used a soil containing microbes likely to have a negative effect against PPNs26.
Here, we developed a novel method to circumvent the use of PPNs under in vitro conditions in order to develop a high-throughput screening method of generalist pathogens against nematodes. Our methodology consisted of creating a soil slurry of microbes and applying it to petri dishes containing the easily-culturable C. elegans. The screening of microbial candidates on C. elegans as opposed to RKNs expedited the process of antagonistic microbe identification. With this method, we were able to rapidly isolate an antagonistic fungus of C. elegans. Subsequently, the fungus was tested and confirmed to be antagonistic against the highly aggressive RKN of potatoes, Meloidogyne chitwoodi both in vitro and in vivo.
Results
Screening of microbes using C. elegans methodology
A total of 16 microbial isolates were recovered from the process of transferring paralyzed and/or dead nematodes to sterile media, post-inoculation of slurries from 3 different soil sources. For more information on the preliminary screening process, see Supplementary Note S1 and Fig. S1. Microbes growing around dead nematodes were determined to be bacterial or fungal based on visual appearances. For instance, if there were biofilm growing around the paralyzed nematode - the nematode was transferred to Luria-Bertani Agar (LBA) as it was assumed to be of bacterial origin. In contrast, if hyphae were actively growing around the paralyzed nematode, the worms were removed from the culture and placed into Potato Dextrose Agar (PDA) to recover the fungus. A challenge brought upon by using this methodology was determining if the nematode’s death was caused by one single microorganism or a combination of microbial action. This issue was overcome by isolating the 16 initial microbial candidates three times in order to obtain pure cultures of the candidate nematophage(s). Organisms were grown at 26 °C for at least 24 hours and once pure broth cultures of all isolates were obtained, the microbial isolates were inoculated onto high population C. elegans plates and monitored for nematode parasitism/antagonism. The inoculation process of pure culture was repeated 3 times for each microbe in order to confirm the successful nematode parasitism/antagonism brought upon by the microbial candidates. After testing all of the isolates independently of one another, only the most effective fungal candidate was identified and investigated further for its nematicidal properties against RKNs (Supplementary Table S1). The fungus was identified as Mortierella globalpina.
Mortierella globalpina expressed the most rapid and effective nematode pathogenesis compared to the other microbes tested in this study. The other fifteen isolates expressed no effect or limited pathogenesis on C. elegans when tested in isolation as compared to the visible immobilization and subsequent nematophage behavior brought upon by M. globalpina (see Supplementary Data for additional information).
In vitro experimentation of Mortierella globalpina on Caenorhabditis elegans
It was determined that C. elegans populations were effectively decimated with the inoculation of M. globalpina spores to the nematode culture. After fungal inoculation, both adult and juvenile nematodes, as well as eggs of C. elegans, were monitored over the course of 72 hours at room temperature (28.8 °C ± 2 °C) until the fungus had completely colonized the petri dish leaving only paralyzed nematodes. It was observed that the fungus fully infected all life stages of C. elegans in vitro in up to 72 hours (Supplementary Table S1). In other words, M. globalpina infected eggs, larval-stages (Larval stage 1 [L1] to L4) and adults (nematodes capable of egg laying) of C. elegans.
Identification of nematophagous fungus
The genetic sequence of the fungal isolate in question was confirmed to be Mortierella globalpina via a matching of the 479 base pairs to a sequence in the public database GeneBank with 100% similarity (NCBI Accession No. NG_064079).
Experimentation of Mortierella globalpina with Meloidogyne chitwoodi
The nematophagous nature of M. globalpina observed against C. elegans was once again observed in vitro during pathogenicity experimentation with M. chitwoodi (Supplementary Table S1). In just 72 hours of exposure to the fungus, the populations of M. chitwoodi nematodes were immobilized by M. globalpina. In two of the five replicate-populations (containing approximately 30 ± 14 nematodes), all nematodes were immobilized. In the remaining three replicates, only two nematodes per replicate survived parasitism by M. globalpina within the first 72 hours (Supplementary Table S1). Over the course of 72 hours it was observed that M. globalpina was pathogenic to both the eggs and juvenile stages (Juvenile stage 2 [J2] – J4), as well as adult nematodes (capable of reproduction) of M. chitwoodi in vitro.
Scanning electron microscope imaging
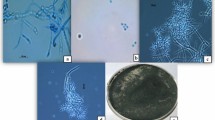
Under the electron microscope, the fungus was confirmed to infect both eggs, larvae, and adults of M. chitwoodi (Fig. 1). It was observed that the fungal mycelium attached and surrounded the eggs of M. chitwoodi. Similar types of attacks were observed on larvae and adults (Fig. 1). In all instances, the mycelium was able to penetrate the cuticle and presumably digest the cellular contents of the nematodes and/or the eggs.
Scanning electron micrographs of Mortierella globalpina pathogenesis on Meloidogyne chitwoodi nematodes. (a) Hyphae of M. globalpina penetrating egg of M. chitwoodi. (b) Higher magnification of hyphae from M. globalpina growing and penetrating egg of M. chitwoodi (c) Hyphae from M. globalpina growing inside M. chitwoodi egg and digesting cuticle layer of M. chitwoodi egg. (d) Higher magnification of hyphae from M. globalpina growing inside M. chitwoodi egg and digesting inner contents of egg. (e) M. globalpina hyphae fully colonizing inner-cuticle of M. chitwoodi adult and presumptively digesting cellular contents. (f) Hyphae of M. globalpina colonizing inner contents of adult M. chitwoodi nematode to then reemerge from nematode cuticle to continue search for nutrition. Six images were combined into one figure with Adobe Photoshop CS6.
The effect of M. globalpina on rutgers var. tomato plant growth parameters
The total plant height as well as fresh and dry weight biomass of tomato was recorded upon harvest. The plants treated with only M. globalpina spores as well as the M. globalpina spores plus M. chitwoodi eggs group expressed significant height increases compared to the control (no applications of M. globalpina nor M. chitwoodi) (p-values: 0.030, 0.003 respectively) (Fig. 2a). Additionally, the treatment inoculated with spores of M. globalpina showed significant fresh-weight increases compared to the control (p-value: 0.005) (Fig. 2b). The M. globalpina spores treatment also expressed a significant increase in dry-weight biomass production compared to the control (p-value: 0.022) (Fig. 2c).
Total height and biomass (fresh and dry) of Rutgers var. tomato plants on harvest day (average of 15 replicates, displayed as mean ± standard deviation). (a) Height. M. globalpina spores and M. globalpina spores plus M. chitwoodi eggs treatments both showed significant height increases compared to the control (+7.13 cm and +9.12 cm, respectively) (p-values: 0.03, and 0.003 respectively, Tukey test). (b) Total fresh-weight biomass of treated Rutgers var. tomato plants on harvest day. M. globalpina spores-inoculated treatment showed significant fresh-weight increases compared to the control (+20.20 g) (p-value: 0.005, Tukey test). (c) Total dry-weight biomass of Rutgers var. tomato plants on harvest day. M. globalpina spores treatment showed significant dry-weight increases compared to the control (+2.83 g) (p-value: 0.02, Tukey test).
Root scanning analysis
Only the roots of the M. chitwoodi egg-inoculated plants and those of the M. globalpina spores plus M. chitwoodi eggs inoculated treatments were subject to root scanning and analysis. This is because all plants subject to nematode infection could be analyzed for root growth abnormalities, whilst the M. globalpina spore-only inoculated plants were used to monitor for any plant growth promoting fungal (PGPF) effects on tomato. Of all parameters analyzed (see Supplementary Table S2), plants inoculated with M. globalpina spores plus M. chitwoodi eggs performed significantly better than M. chitwoodi eggs only treatment. The spores plus eggs treatment resulted in increased number of root-tips (Fig. 3a) and reduced number of root-galls (Fig. 3b) compared to the M. chitwoodi eggs only inoculated treatment (p-values: 0.0178, and 1.19e−10 respectively). In this study, the reduced number of nematode infection sites (galls) within tomato roots was an indicator of effective biocontrol of RKNs.
WinRHIZO Root scan analysis (average of 15 replicates, displayed as mean ± standard deviation): (a) Root Tips. M. globalpina spores plus M. chitwoodi eggs-inoculated plants significantly increased number of root-tips compared to M. chitwoodi eggs-only inoculated treatment (+1800.4 root tips) (p-value = 0.018, Tukey test). (b) Root Galls. Root scan analysis of number of root galls (gall = PPN infection site). M. globalpina spores plus M. chitwoodi eggs-inoculated treatment significantly decreased the number of root galls compared M. chitwoodi eggs-only inoculated treatment (−83.8 galls) (p-value: 1.19e−10, Tukey test).
Discussion
In recent years, chemicals like 1,3-dichloropropene (1,3-D) and metam sodium have been utilized as alternatives to the use of methyl bromide27. It was found that the most promising chemical alternatives to methyl bromide for PPN control were the combinations of 1,3-D-chloropicrin, chloropicrin-proprietary solvent, and 1,3-D-metam sodium27. However, compounds like metam sodium can have adverse ecological and human health effects28. Pruett et al. suggests that metam sodium can induce both allergic dermatitis and/or asthma in humans28. Thus, ecologically friendly alternative methods to control PPN (such as beneficial microbes) are needed.
There is palpable need to identify new microbes that could control plant parasitic nematodes of economic importance29. However, certain difficulties arise in the search for new nematophagous microorganisms such as the lack of a widely used pathogenicity-screening method. In this study one of our aims was to develop a simple and high-throughput methodology to identify new beneficial microbes that could serve as general-antagonists against nematodes. Although ideally any screening would be target specific, the difficulty in culturing PPNs in vitro makes this unreasonable. This difficulty has also resulted in chemical nematicides that are non-specific. However, by using a free-living nematode model, we reasoned that we could develop a targeted approach that is limited to nematodes. Both PPNs and free-living nematodes share the same soil habitat and are subject to similar pressures such as the presence of pathogens. Accordingly, it has been shown that different types of soil nematodes were affected similarly when mammalian grazing was introduced to the above-ground land of their soil habitat30. Similarly, nematodes of different orders experienced analogous population decline or elimination as a result of increasing heavy metal contents in an agroecosystem soil31. These studies contribute to the assumption that nematodes of different genera are susceptible to similar stresses.
We used the soil microbiome as a natural reservoir of potential microbial biocontrol agents against PPN. We specifically selected a soil (Sargent farm) that has been previously analyzed in terms of microbiology and showed low ratios of PPN26. The total nematode faunal profile in the Sargent farm soil proposed by Castillo et al. was 10% Herbivores, 62% bacterivores and 28% fungivores. Based on this nematode profile, the soils of the Sargent farm fit into ‘Quadrat D’ of the soil food web proposed by Ferris et al.26,32. Characteristics of the ‘Quadrat D’ show a food web that is generally degraded and includes both a high C:N ratio and fungi being the primary driver of decomposition32. With the Sargent farm soil expressing a low population of herbivorous nematodes as well as fungi being the primary source of decomposition, we hypothesized that this soil was likely to contain a fungal community pathogenic to PPNs.
Caenorhabditis elegans was used as a model organism in our study because it is easily established and commonly cultured under in vitro conditions33. In further support of the use of C. elegans as a model organism, the nematode has also been used as a model in other studies such as high-throughput drug discovery for anti-microbials, anti-infectives, immunity responses, host-pathogen interactions, and several other bioactive responses34,35,36,37. Interestingly, Hsueh et al. utilized C. elegans to characterize the attraction and trapping ability of the fungus Arthrobotrys oligospora38. However, the fungus A. oligospora had been characterized previously by Fresenius in 1850 and C. elegans was merely used to visualize and photograph the nematophage in action38,39.
In our study, the fungus M. globalpina was selected as the candidate for experimentation as it was observed to parasitize eggs, larvae and adults of C. elegans nematodes. Additionally, M. globalpina also expressed effective nematode pathogenicity in its isolated form rather than in combination with other microbes. There is little information about the members of the fungal genus Mortierella serving as nematophagous fungi. Orion et al. reported that Mortierella spp. was observed inside the gelatinous matrix of RKN egg masses40. Mortierella spp. hyphae were also observed in hatched eggs of RKN, suggesting a nematophagous nature by one or several fungi of the Mortierella genus40. However, the member Mortierella alpina was tested in greenhouse studies against M. javanica, but was most effective when co-inoculated with Trichoderma longibrachiatum41.
Our results suggest that M. globalpina produces extracellular enzymes that can degrade the cuticle of both C. elegans and M. chitwoodi. It is worth noting the cuticle of most nematodes is largely composed of collagen24; thus, it is likely that M. globalpina might produce and secrete collagenases. This report is to the best of our knowledge the first time that M. globalpina has been introduced as a biocontrol agent of RKNs. Additionally, the application of M. globalpina spores alone increased root and shoot biomass of Rutgers var. tomato plants compared to other treatments. This suggests that M. globalpina may have some plant growth promoting effects on tomato. Although not tested here, Mortierella antarctica and Mortierella verticillata have been observed to synthesize gibberellic acid and indoleacetic acid in vitro, which may explain the growth promoting effects on tomato of M. globalpina42.
In conclusion, this study reports a new method to rapidly identify generalist nematode pathogens by utilizing the free-living C. elegans as a model. In addition, we state that a newly-isolated nematophagous Mortierella globalpina reduced nematode infection sites (galls) and could potentially serve as a useful biocontrol agent against RKNs in cropping systems. However, further field studies are necessary to determine (i) the usefulness of M. globalpina as a biocontrol measure on other herbivorous nematodes (non-RKNs), (ii) the ecological impacts on the natural soil microbiota when using M. globalpina as a biocontrol agent, (iii) the effect of M. globalpina inoculation as a protective measure in known PPN/RKN-infested soils, and (iv) the mechanism of chemical action for the pathogenesis of M. globalpina against M. chitwoodi.
Methods
Experimentation with Caenorhabditis elegans
Soil slurry preparation and inoculation
In screening for nematophagous microbes we used the ‘Sargent’ potato field soil listed in Castillo et al., which has a strong likelihood of containing beneficial microbes26. A two-part ‘Sargent’ soil and one-part molecular water slurry was prepared. This slurry functioned as a liquid suspension of microorganisms present within the Sargent field soil. The water and soil mixtures were contained in a 50 ml VWR Falcon tube and were homogenized for 2 hours on a Fisher Vortex Genie at maximum speed. The slurry was then centrifuged at 2440 RPM in a Sorvall Super T21 benchtop centrifuge for 10 minutes at room temperature (28.8 °C ± 2 °C) to pellet any remaining soil debris. Subsequently, the slurry was aspirated from the Falcon tube, and serially diluted to prepare 10−6, 10−7 and 10−8 dilutions. Aliquots of 50 ul and 100 ul of each slurry dilution were plated onto high population (>1,000) C. elegans culture plates (described in 4.1.2) to monitor for microbial pathogenesis (Supplementary Fig. S1). The studies were repeated three times using three replicates for each solution/dilution in order to locate as many nematophagous microbial candidates as possible. For a full description of this methodology and soils tested in preliminary experimentation, see Supplementary Note S1.
Population establishment of Caenorhabditis elegans
Caenorhabditis elegans nematodes were obtained from the Biochemistry Department at Colorado State University in Fort Collins, CO. Nematodes were cultured in vitro on 9 cm petri dishes prepared with Nematode Growth Media (NGM) agar33. Nematode growth media agar consists of: 3 g NaCl, 2.5 g bacterial peptone, 17 g agar and 975 ml of diH2O. After autoclaving, the following compounds are mixed into the NGM: 1 ml of 1 M CaCl2, 1 ml of 1 M MgSO4, 25 ml of 1 M KPO4 and 1 ml cholesterol in ethanol (5 mg/ml). To provide a food source for C. elegans, Escherichia coli OP50 was grown onto Luria Bertani agar (LBA) at 37 °C until a bacterial lawn was established. Once the lawn was formed, an 8 mm by 8 mm square chunk of E. coli OP50 was excised from the LBA and seeded onto the NGM agar. The seeded NGM was incubated at 37 °C until the E. coli formed a new bacterial lawn. After bacterial establishment, the petri dishes were inoculated with 30 ul suspension of C. elegans (containing 10 ± 4 eggs or stage 1 larvae (L1) of bacterivorous nematodes) obtained using a common nematode synchronization methodology33.
Determining microbial pathogenicity against Caenorhabditis elegans
Caenorhabditis elegans nematodes assumed to be antagonized by nematophagous microorganisms were examined for behavioral difficulties such as lack of movement and/or nematode body stiffening. Those nematodes expressing damaged cuticle layers, and/or presence of bacteria [biofilm] or fungi [hyphae] around the body were identified. The nematodes that were infected were removed from the NGM via the use of a needle from an Echinopsis pachanoi cactus mounted with parafilm to a wooden stirrer. Once located, the affected nematodes were placed onto either Potato Dextrose Agar (PDA) for fungal isolates or Luria Bertani Agar (LBA) for bacterial isolates to subsequently isolate individual colonies. Isolates were repeatedly sub-cultured, at least 3 times, until a pure culture was obtained and used for further experimentation. Bacterial isolates were incubated in the dark at 37 °C and fungal isolates were incubated in the dark at room temperature (28.8 °C ± 2 °C).
Confirmation of microbial-pathogenicity against C. elegans
After obtaining pure cultures from the isolates assumed to be antagonistic or pathogenic to C. elegans, the microbes were cultured into a broth (liquid media, PD broth or LB broth). The liquid isolates were grown to a concentration of 1 × 108 spores or cells/ml and then inoculated onto C. elegans plates. Inoculated plates were examined over the course of 72 hours in order to monitor and confirm any antagonism or pathogenic attacks against C. elegans in vitro. Microbial pathogenicity confirmation was repeated in triplicate prior to testing the isolated organism(s) against Melodogyne chitwoodi.
Experimentation with Meloidogyne chitwoodi
In Vitro experimentation
Rutgers var. tomato seeds were surface sterilized with a 2.5% sodium hypochlorite (NaClO) solution, rinsed 5 times with sterile water and sown into water-agar (10 grams per liter, pH 7.4) at four seeds per petri dish (See Supplementary Image S1). After one week submerged in the agar, when the length of the roots ranged from 1 cm-2 cm long, in vitro experimentation began. Fungal spores were cultured in liquid (Potato dextrose broth) and were inoculated onto the agar surrounding tomato roots at 20 uL/root at a concentration of 1 × 108 spores/ml. Additionally, 30 ± 14 M. chitwoodi eggs were then added in the center of the 9 mm petri dish to examine effects of the fungal isolate as a biocontrol agent against M. chitwoodi nematodes. Five replicates of this experiment were prepared for testing, and the experiments were repeated in triplicate to confirm results. Petri dishes were monitored for 72 hours to examine signs of plant parasitism by nematode, or nematode pathogenesis by M. globalpina. Observations of infected or immobilized nematodes were recorded until all present nematodes were infected by the fungus (~72 h).
Identification of nematophagous fungal isolate
A pure culture of a nematophagous fungus with pathogenic activity against both C. elegans and M. chitwoodi was cultured on sterile PDA and grown for 24 hours in the dark at room temperature (28.8 °C ± 2 °C). Freshly grown samples were then sent to MIDI labs (Newark, DE) where the 28s large subunit (LSU) rRNA gene sequencing was conducted in order to obtain the organism’s identity. The protocol to generate the 28s rRNA gene sequence is as follows: Approximately 300 base pairs of the variable D2 region of the LSU rRNA gene is PCR amplified from genomic fungal DNA. The primers used for the PCR and sequencing correspond to positions 3334 F and 3630 R in the Schizosaccharomyces japonicus LSU rRNA gene. Amplification products are purified from excess primers and dNTPs and checked for quality and quantity by running a portion of the products on an agarose gel. Cycle sequencing of the 16S rRNA amplification products were carried out using DNA polymerase and dye terminator chemistry. Excess dye-labeled terminators were then removed from the sequencing reactions. The samples were electrophoresed on either a 3,130 or 3,130xl Genetic Analyzer
In Vivo experimentation
Rutgers var. Tomato seedlings were pre-germinated in Promix Bx peat moss and grown in plug trays until two sets of true leaves had emerged on the seedlings. This experiment consisted of three treated groups of tomato plants and a control, and all groups consisted of 15 replicates. Treatments were determined in order to observe the effect of M. globalpina on tomato plants infected with PPN. The treatments were: M. chitwoodi eggs, M. chitwoodi eggs plus M. globalpina spores, and M. globalpina spores alone, and control group. In all treatments; roots of tomato seedlings were washed of adhering substrate and suspended in either M. globalpina spores cultivated in PDB at [1 × 108 spores/ml], sterile PDB or sterile water for 20 minutes41. After root-dip inoculation, plants were transplanted into a (1:1:1) Promix Bx: Vermiculite: Sand substrate to allow for effective nematode movement throughout. After transplanting tomato seedlings, M. chitwoodi eggs were added at 10,000 eggs/plant. The eggs were inoculated into the substrate at the soil level near the root zone by the methodology described by AL-Shammari et al.41. Plants that underwent fungal-root-dip inoculation (but were not infected with eggs of M. chitwoodi) were carried out in order to monitor for any plant growth promoting fungal effects on tomato. The experiment was conducted in CSU’s Horticulture Center Greenhouse and carried out in a randomized block design provided by random.org, with each plant pot serving as a block. This in vivo experiment was repeated 5 times in order to confirm results. Plants were grown for a total of 7 weeks and harvested 5 weeks after the nematode inoculation. Upon harvesting, fresh and dry weight biomass (above-, below-ground, and total) were recorded, along with the final plant height. Additionally, the roots of the plants exposed to M. chitwoodi eggs were washed of all substrate prior to biomass recording and were scanned and analyzed by WinRHIZO Pro software. After analysis of roots, all plant matter was oven dried at 70 °C for until there was no change in dry weight biomass measurement.
Root scanning and analysis
Only roots of plants treated with M. chitwoodi eggs were scanned for analysis. Tomato plants were uprooted on harvest day and roots were washed free of any adhering substrate. Roots were then removed from shoots, submerged into shallow trays of water and scanned using an EPSON Perfection V750 Pro Photo Scanner. Once the images of roots were acquired, they were analyzed by Reagent Instrument Inc.’s WinRHIZO Pro. The total root length, surface area, average root diameter, root volume, number of crossings, the number of root galls, and the number of root forks were observed and analyzed via the use of WinRHIZO Pro analysis software. In our study, number of galls were used as a measure to show effectiveness (or lack thereof) of M. globalpina in controlling RKNs, and reduced symptomology of nematode infection (or a reduction in the number of root-galls) was an indicator of biocontrol agent effectiveness.
Electron microscope imaging
The fungus M. globalpina was grown on 5 cm petri dishes filled with water-agar (pH 7.4) and planted with four Rutgers var. tomato seeds similar to the methodology in 4.2.1, but smaller (5 cm) petri dishes. Once the mycelium began to grow onto the tomato roots; eggs and L1 larvae of M. chitwoodi were added to the center of the water agar dishes. The petri dishes were sealed with parafilm and placed in the dark at 26 °C. Plates were examined over the course of 26 hours and any fungal attacks on the nematodes or eggs were excised from the agarose plates and stored in a glutaraldehyde fixative (pH 7.4) at 4 °C for later SEM processing. For SEM success, the samples needed to be 1 mm3 and were prepared with a No. 10 scalpel blade under a laminar flow hood. The samples were then processed through critical point drying, mounting, and subsequently imaged on a Scanning Electron Microscope at Colorado State University’s Central Instrument Facility. Samples were prepared in 50 mm agar petri dish with fungus, nematodes and newly germinated tomato plant roots.
Statistical analysis
The differences between groups in all results were analyzed by one-way ANOVA and if significant, were further analyzed with Tukey’s HSD test with R statistical analysis software. Figures showing significance in both plant growth parameter and root scan results were created with R statistical software.
Data Availability
The datasets generated during and/or analyzed throughout the current study are available from the corresponding author upon reasonable request.
References
Wang, D. Y., Kumar, S. & Hedges, B. S. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proceedings of the Royal Society of London B: Biological Sciences, 163–171 (1999).
Yeates, G. W., Bongers, T., De Goede, R. M., Freckman, D. W. & Georgieva, S. S. Feeding habits in soil nematode families and genera - an outline for soil ecologists. The Journal of Nematology, 315–331 (1993).
Nicol, J. M. et al. Current nematode threats to world agriculture. Genomics and Molecular Genetics of Plant-Nematode Interactions (2011).
Karajeh, M. R. Checklist of host range of root-knot nematodes (Meloidogyne species and races) in Jordan. Jordan Journal of Agricultural Sciences 11(3), 761–769 (2015).
Quentin, M., Abad, P. & Favery, B. Plant parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Frontiers in Plant Science 4, 1–7 (2013).
Sasser, J. N. & Freckman, D. W. A world prospective on nematology: the role of the society. Vistas on Nematology 2(3), 7–14 (1987).
Brown, R. & Kerry, B. Principles and practice of nematode control in crops. Academic Press, 1–12 (1987).
Nolin, J. W. & Becker, J. O. The challenge of research and extension to define and implement alternatives to methyl bromide. The Journal of Nematology 26(4s), 573–586 (1994).
Saucet, S. B., Ghelder, C. V., Abad, P., Duval, H. & Esmenjaud, D. Resistance to root-knot nematodes Meloidogyne spp. in woody plants. New Phytologist 211(1), 41–56, https://doi.org/10.1111/nph.13933 (2016).
Djian-Caporalino, C. et al. Root-knot nematode (Meloidogyne spp.) Me resistance genes in pepper (Capsicum annuum L.) are clustered on the P9 chromosome. Theoretical and Applied Genetics 114(3), 473–486 (2007).
Kokalis-Burelle, N. Pasteuria penetrans for control of Meloidogyne incognita on tomato and cucumber, and M. arenaria on snapdragon. Journal of Nematology 47(3), 207–213 (2015).
Oka, Y., Shapira, N. & Fine, P. Control of root-knot nematodes in organic farming systems by organic amendments and soil solarization. Crop Protection 26(10), 1556–1565 (2007).
Kiewnick, S. & Sikora, R. A. Biological control of the root-knot nematode Meloidogyne incognita by Paecilomyces lilacinus strain 251. Biological Control 38(2), 179–187 (2006).
Sayre, R. M., Wergin, W. P., Schmidt, J. M. & Starr, M. P. Pasteuria nishizawae sp. nov., a mycelial and endospore-forming bacterium parasitic on cyst nematodes of genera Heterodera and Globodera. Research in Microbiology 142(5), 551–564 (1991).
U.S., Environmental Protection Agency, Gagliardi, J. V., Kough, J. L., Borges, S., & Tomimatsu, G. Biopesticided Registration Action Document: Paecilomyces lilacinus strain 251. (2005).
U.S., Environmental Protection Agency, Gagliardi, J. V., Kough, J. L., Borges, S., & Tomimatsu, G. Biopesticides registration action document: Pasteuria nishizawae – Pn1. (2005).
Dong, L. W. & Zhang, K. Q. Microbial control of plant-parasitic nematodes: a five-party interaction. Plant Soil 288(1), 31–45, https://doi.org/10.1007/s11104-006-9009-3 (2006).
Diaz-Manzano, F. E. et al. Long-term in vitro system for maintenance and amplification of root-knot nematodes in Cucumis sativus roots. Frontiers in Plant Science, 7, https://doi.org/10.3389/fpls.2016.00124 (2016).
Breger, J. et al. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLOS Pathog. 3(2), e18, https://doi.org/10.1371/journal.ppat.0030018 (2007).
Moy, T. I. et al. Identification of novel antimicrobials using a live-animal infection model. Front in Microbiology 6, 1280 (2006).
Sifri, C., Begun, J., Ausubel, F. M. & Calderwood, S. B. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infection and Immunity 7(4), 2208–2217 (2003).
Muhammed, M., Coleman, J. J. & Mylonakis, E. Caenorhabditis elegans: a nematode infection model for pathogenic fungi. Methods in Molecular Biology, 447–454 (2012).
Sem, X., & Rhen, M. Pathogenicity of Salmonella enterica in Caenorhabditis elegans relies on disseminated oxidative stress in the infected host. PLOS One. 7(9), https://doi.org/10.1371/journal.pone.0045417 (2012).
Davies, K. G. & Curtis, R. H. Cuticle surface coat of plant-parasitic nematodes. Plant Pathology and Microbiology 49, 135–156 (2011).
Kearn, J., Ludlow, E., Dillon, J., O’Conner, V. & Holden-Dye, L. Fluensulfone is a nematicide with a mode of action distinct from anticholinesterases and macrocyclic lactones. Pesticide Biochemistry and Physiology 109, 44–57 (2014).
Castillo, J. D., Vivanco, J. M. & Manter, D. K. Bacterial microbiome and nematode occurance in different potato agricultural soils. Microbial Ecology 74(4), 888–900 (2017).
Desaeger, J., Dickson, W. D. & Locascio, S. J. Methyl bromide alternatives for control of root-knot nematode (Meloidogyne spp.) in tomato production in Florida. The Journal of Nematologists 49(2), 140–149 (2017).
Pruett, S. B., Myers, L. P. & Keil, D. E. Toxicology of metam sodium. Journal of Toxicology and Environmental Helath 4(2), 207–222 (2001).
Kaplan, J. K. The USDA Nematode Collection. Retrieved from United States Department of Agriculture AgResearch Magazine: https://agresearchmag.ars.usda.gov/2016/may/nematode/ (2016)
Andriuzzi, W. S. & Wall, D. H. Grazing and resource availability control soil nematode body size and abundance–mass relationship in semi arid grassland. Journal of Animal Ecology, 87 ( 5 ), (2018).
Georgieva, S. S., McGrath, S. P., Hooper, D. J. & Chambers, B. S. Nematode communities under stress: the long-term effects of heavy metals in soil treated with sewage sludge. Applied Soil Ecology 20, 27–42 (2002).
Ferris, H., Bongers, T. & de Goede, R. A framework for soil food web diagnostics: extension of the nematode faunal analysis concept. Applied Soil Ecology 18, 13–29 (2001).
Eisenmann, D. M. The C. elegans Research Community, https://doi.org/10.1895/wormbook.1.7.1 (2005).
Aballay, A. & Ausubel, F. M. Caenorhabditis elegans as a host for the study of host–pathogen interactions. Current Opinion in Microbiology 5, 97–101 (2002).
Desalermos, A., Muhammed, M., Glavis-Bloom, J. & Mylonakis, E. Using C. elegans for antimicrobial drug discovery. Expert Opinion on Drug Discovery 6(6), 645–652 (2012).
Marsh, E. K. & May, R. C. Caenorhabditis elegans, a model organism for investigating immunity. Applied and Environmental Microbiology 78(7), 2075–2081 (2012).
O’Reilly, L. P., Luke, C. J., Perlmutter, D. H., Silverman, G. A., & Pak, S. C. C. elegans in high-throughput drug discovery. Advanced Drug Delivery Reviews, 247–253 (2015).
Hsueh, Y. et al. Nematophagous fungus Arthrobotrys oligospora mimics olfactory cues of sex and food to lure its nematode prey. eLIFE. https://doi.org/10.7554/eLife.20023.001 (2017).
Fresenius, G., Frankfurt, A. (ed.) Beiträge zur Mykologie (1850).
Orion, D., Kritzman, G., Meyer, S. L., Erbe, E. F. & Chitwood, D. J. A role of the gelatinous matrix in the resistance of root-knot nematode (Meloidogyne spp.) eggs to microorganisms. The Journal of Nematology 33(4), 203–207 (2001).
AL-Shammari, T. A., Bahkali, A., Elgorban, A., Elkahky, M. & Al-Sum, B. The use of Trichoderma longibrachiatum and Mortierella alpina against root-knot nematode, Meloidogyne javanica on tomato. Journal of Pure and Applied Microbiology 7, 199–207 (2013).
Ozimek, E. et al. Synthesis of indoleacetic acid, gibberellic acid and 3 ACC-deaminase by Mortierella strains promote 4 winter wheat seedlings growth under different 5 conditions. Preprints, https://doi.org/10.20944/preprints201809.0298.v1.
Acknowledgements
This research was supported by Colorado State Universities Agricultural Experiment Station. We would like to thank both present and past members of the Vivanco lab for fruitful comments during the composition of this manuscript.
Author information
Authors and Affiliations
Contributions
J.V. developed the main idea for this experiment. M.D. conducted the experiment under the guidance of J.V. M.D. wrote the manuscript with revisions provided by J.V. and D.M. M.D. conducted statistical analyses of the data in this experiment and prepared all images, figures and Supplementary Information for the results of this study. All authors reviewed the manuscript prior to submission.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
DiLegge, M.J., Manter, D.K. & Vivanco, J.M. A novel approach to determine generalist nematophagous microbes reveals Mortierella globalpina as a new biocontrol agent against Meloidogyne spp. nematodes. Sci Rep 9, 7521 (2019). https://doi.org/10.1038/s41598-019-44010-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-44010-y
This article is cited by
-
Microbial antagonists: diversity, formulation and applications for management of pest–pathogens
Egyptian Journal of Biological Pest Control (2023)
-
Combined application of microbial inoculant and kelp-soaking wastewater promotes wheat seedlings growth and improves structural diversity of rhizosphere microbial community
Scientific Reports (2023)
-
Prebiotics: A Solution for Improving Plant Growth, Soil Health, and Carbon Sequestration?
Journal of Soil Science and Plant Nutrition (2023)
-
Genetic disruption of Arabidopsis secondary metabolite synthesis leads to microbiome-mediated modulation of nematode invasion
The ISME Journal (2022)
-
Intercropping with marigold promotes soil health and microbial structure to assist in mitigating tobacco bacterial wilt
Journal of Plant Pathology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.