Abstract
Aphids produce wing and wingless morphs, depending on the environmental conditions during their complex life cycles. Wing and wingless variations play an important role in migration and host alternation, affecting the migration and host alternation processes. Several transcriptional studies have concentrated on aphids and sought to determine how an organism perceives environmental cues and responds in a plastic manner, but the underlying mechanisms have remained unclear. Therefore, to better understand the molecular mechanisms underlying the wing polyphenism of this fascinating phenomenon, we provide the first report concerning the wing development of aphids in bird cherry-oat aphid Rhopalosiphum padi with comparative transcriptional analysis of all the developmental stages by RNA-Seq. We identified several candidate genes related to biogenic amines and hormones that may be specifically involved in wing development. Moreover, we found that the third instar stage might be a critical stage for visibility of alternative morphs as well as changes in the expression of thirty-three genes associated with wing development. Several genes, i.e., Wnt2, Fng, Uba1, Hh, Foxo, Dpp, Brk, Ap, Dll, Hth, Tsh, Nub, Scr, Antp, Ubx, Asc, Srf and Fl, had different expression levels in different developmental stages and may play important roles in regulating wing polyphenism.
Similar content being viewed by others
Introduction
Organisms can flexibly alter their phenotypes in response to external stimuli to adapt to their surrounding environments, a phenomenon referred to as phenotypic plasticity1. In particular, some species can display different morphs of phenotypic plasticity despite having the same genotype in response to specific environments, a phenomenon referred to as polyphenism2,3. This is an adaptive behavioral strategy widely used by insects that live in heterogeneous environments, such as the wing and wingless morph development of aphids4,5,6.
Aphids are very important sap-sucking insects in Aphidoidea (Hemiptera), with more than 5000 known species in the world7. Aphids with complicated and diverse biotypes can reproduce wing or wingless morphs by distinct morphological differentiation in some species in response to environmental changes8. Wing and wingless variation is one of the normal characteristics to most aphid species; but wing morph variation is even more important to aphids with the complex and diverse life cycle, for example, the winged morphs of aphids are adapted for flight and migration to new locations, and the host-alternating species are wing-dimorphic, making it possible to alternate between primary hosts and secondary hosts to complete their life cycle. Serious pest challenges could then occur in any of the migration and host alternation processes. Therefore, wing and wingless variation is not only a trade-off strategy that has evolved in most insect orders for coping with complex and uncertain environments but also a key factor that leads to population expansion and infestation9,10,11.
Earlier studies have been based on morphological characters or transcriptional data and sought to determine how an organism perceives environmental cues and responds in a plastic manner, but the underlying mechanisms have remained unclear10,12,13,14,15,16,17. Utilizing scanning electron microscopy and histological sectioning, Ishikawa et al.10 observed and compared wing development in the winged and wingless individuals in Acyrthosiphon pisum and found the developmental processes underlying wing polyphenism10. Previous transcriptional studies have focused on some developmental stages of aphids, e.g., winged and wingless adults12,14,16,17, embryo and winged and wingless adults15, or fourth instar nymphs and winged and wingless adults13 and identified a handful of individual winged-associated genes, functional GO terms and pathways.
The bird cherry-oat aphid Rhopalosiphum padi (Linnaeus, 1758) is found in Rhopalosiphum, Aphidini (Aphididae: Aphidinae) and is distributed virtually worldwide, including in China, Japan, the Korean Peninsula, America, Canada, New Zealand, Russia, Egypt, Jordan, and Europe18. R. padi is holocyclic and anholocyclic, and is conservative in its choice of primary host, which includes Armeniaca, Amygdalus, Malus, Pyrus and Prunus plants. R. padi can alternate to secondary hosts, such as Cyperaceae, Iridaceae, Juncaceae, Typhaceae, dicots and Tuphaceae plants18,19,20. Therefore, R. padi is a typical wing-dimorphic aphid and its clonal populations, with the same genetic backgrounds, can be easily reared in the laboratory. Thus, R. padi is a useful experimental model for studying wing polyphenism.
The molecular mechanisms underlying the induction of winged and wingless aphids remain unknown. To better understand the gene regulatory basis of this fascinating phenomenon, we provide the first report concerning the wing development of R. padi with comparative transcriptional analysis of all developmental stages by RNA-Seq. Our results provide significant insights into the molecular mechanisms underlying how an organism relays environmental signals to its instars, increases our understanding of the critical stage that triggers alternative morph determination and provides a guide for monitoring the dynamics of migratory incidence in pests, predicting outbreaks of hazardous pests, and determining methods to control them.
Results
Offspring produced with winged morphs
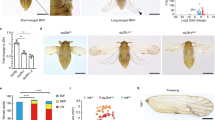
Parthenogenetic females were subjected to crowded or solitary conditions, and we found that there were no obvious differences in external morphology in the first instar and second instar stage nymphs. In the third instar stage, small wing buds were identified in winged aphids, while there were no swollen structures in the thoracic part of the wingless aphids. Furthermore, during the fourth instar and adult stages, wing buds were enlarged and matured in winged aphids, while wing primordia completely disappeared in wingless aphids. In addition, we collected individuals for each instar aphid of the same genotype. Our analysis showed that 90% to 100% of offspring were winged when mothers were reared under crowded conditions for 16 h, whereas parthenogenetic females reared under solitary conditions produced less than 10% winged offspring (Table S1).
RNA-Seq data analysis
The reference gene library was constructed by mixing equal quantities of total RNA from all samples, generating approximately 55.51 million total raw reads for the reference gene set. After filtering out the low-quality reads, we obtained 43.79 million clean reads. After filtering out repetitive and unannotated genes, the merged datasets yielded a total of 39,328 unigenes that were subjected to further analysis, and the total length, average length, N50, N90 and GC content of unigenes are 44,957,242 bp, 1,143 bp, 2,260 bp, 408 bp and 34.38%, respectively (Table S2).
In addition, 48 sample libraries of R. padi were generated, with an average of 22,886,759 raw sequencing reads and 22,741,712 clean reads after filtering low-quality reads (Table S3). After filtering, the clean reads were mapped to the reference gene library using the Bowtie2 tool21. The average mapping ratio with the reference genes was 94.79% (Table S4).
Differentially expressed genes and pathways related to winged morph regulation
We compared the transcriptomes of all instar offspring at five sequential developmental stages based on gene expression results. Many differentially expressed genes and pathways were identified that may be involved in morphological divergence in aphid wing development.
We performed differentially expressed genes analysis and found that several biogenic amines and hormones may play important roles in regulating aspects of wing-morph development. More detailed inspection of the biogenic amines from RNA-Seq data revealed that these biogenic amine effects were mainly associated with serotonin, dopamine and octopamine, including Serotonin Transporter (SeT), Serotonin Receptor 1 (SeR1), Dopamine Transporter (DoT), Dopamine Receptor (DoR, DoR1, DoR2), Octopamine Receptor (OcR, OcR1, OcR2, OcR3). For hormones, such as juvenile hormone, ecdysone and insulin, these were associated with Juvenile Hormone Esterase (JHE), Juvenile Hormone Epoxide Hydrolase (JHEH), Juvenile Hormone Binding Protein (JHBP), Juvenile Hormone Acid Methyltransferase (JHAM), Ecdysone Receptor (EcR), Ecdysone-induced Protein (EcP74, EcP75, EcP78, EcP93), Insulin Receptor (InR), and Insulin Receptor Substrate (IRS).
Moreover, we found thirty-three differentially expressed genes: Flightin (Fl), Wingless (Wg), Engrailed (En), Vestigial (Vg), Apterous (Ap), Hedgehog (Hh), Aristaless (Al), Serrate (Ser), Patched (Ptc), Brinker (Brk), Teashirt (Tsh), Nubbin (Nub), Fringe (Fng), Rhomboid (Rho), Ultrabithorax (Ubx), Homothorax (Hth), Antennapedia (Antp), Decapentaplegic (Dpp), Spalt, Notch, Wnt1, Wnt2, Wnt11, Wnt16, Distal-less (Dll), Cubitus interruptus (Ci), Optomotor-blind (Omb), Sex combs reduced (Scr), Forkhead box protein O (Foxo), Achaete-scute complex (Asc), Serum response factor (Srf), Cell division control protein 42 (Cdc42), and Ubiquitin-activating enzyme E1 (Uba1).
To further identify the functions of these genes, we found that the Wnt, Wingless (Wg), Notch, Hedgehog (Hh), Decapentaplegic (Dpp), Ecdysone, Insulin, and Epidermal growth factor receptor (EGF-R) signaling pathways were coordinately involved in aphid wing development. Wnt signaling pathway regulation included Wnt1, Wnt2, Wnt11, and Wnt16. Wg signaling pathway regulation included Wg and Vg. Notch signaling pathway regulation included Notch, Ser, and Fng. Hh signaling pathway regulation included Hh, Ptc and Ci. Dpp signaling pathway regulation included Dpp, Brk, Omb and Spalt. Ecdysone signaling pathway regulation included EcR, EcP74, EcP75, EcP78, and EcP93. Insulin signaling pathway regulation included InR and IRS. EGF-R signaling pathway regulation included En.
Biogenic amine and hormone-associated gene expression profiling among offspring in crowded and solitary conditions
From the RNA-Seq data, we examined the expression levels of genes associated with the transport and reception of serotonin, dopamine and octopamine and the enzymatic functions, proteins or transport and the reception of juvenile hormone, ecdysone and insulin. We clustered these expression patterns across offspring based on the fragments per kilobases of transcripts per million mapped fragments (FPKM) expression results (Table S5) for crowded and solitary conditions using hierarchical clustering analysis by MeV4.9.022.
The results showed that biogenic amines and hormone-associated genes were grouped based on their expression profiles across all instar samples into 5 manually defined clusters (Fig. 1). The results were quite striking: the biogenic amines associated with serotonin, dopamine and octopamine exhibited similar expression patterns in crowded and solitary conditions. For example, serotonin associated with SeT and SeR1 exhibited a similar trend of expression: the first to third instars showed lower expression values, and the fourth instar to adult stages exhibited a gradually increased trend of higher expression. Dopamine related to DoT, DoR, DoR1, and DoR2 showed lower expression values for all instars. Octopamine showed a similar expression pattern of OcR, OcR1, OcR2, and OcR3, namely, the first instars, which showed a higher expression level, the second and third instars displayed a similar trend of lower expression levels, and the fourth instar to adult stages showed higher expression. Particularly, OcR1 and OcR3 displayed higher expression, while OcR and OcR2 exhibited lower expression.
Heat map of biogenic amine and hormone-associated gene expression profiling among offspring in crowded and solitary conditions by RNA-seq. The FPKM represents the average of three replicates of gene expression levels in each instar aphids. The colored barcode gradients indicate the minimum value in green and the maximum value in red.
Moreover, the hormones related to the juvenile hormone, ecdysone and insulin, also showed a similar trend of expression patterns in crowded and solitary conditions. For instance, juvenile hormones associated with JHE, JHEH, JHBP and JHAM, showed different trends of expression patterns. JHE displayed lower expression levels in the first to fourth instars, while adult stages displayed higher expression values. JHEH showed higher expression values for all samples. JHBP exhibited lower expression levels in the first, second and adult stages and exhibited a gradually increased trend of higher expression levels in the third instar to fourth instar. JHAM showed higher expression values for all the instars, especially in the third instar and fourth instar. Ecdysone exhibited a variety trends in terms of EcR, EcP74, EcP75, EcP78 and EcP93. EcR showed higher expression in the adult stages relative to other instars. EcP74 displayed higher expression levels in the first, second and adult stages and showed slightly lower expression levels in the third instar and fourth instar. In addition, EcP78 showed higher expression values in the fourth instar and adult stages than the others, and EcP93 exhibited higher expression in the third instar to adult winged instar, while EcP75 showed the highest expression values at all aphid stages. Additionally, insulin associated with InR exhibited higher expression levels in the adult stages relative to the other instars, while insulin related to IRS showed higher expression levels at all stages.
Wing-associated gene expression profiling among offspring in crowded and solitary conditions
We examined the transcriptomes of whole-body tissues of aphids at five sequential developmental stages and obtained thirty-three differentially expressed wing-morph regulatory genes. Then, we clustered these thirty-three wing-associated gene expression patterns across all developmental stages based on the FPKM expression results (Table S6) for crowded and solitary conditions.
The results showed that wing-associated genes were grouped by their expression profiles across all samples into seven manually defined clusters (Fig. 2). Upon further inspection of the heat map of the wing-associated gene expression profiling, we found that these genes exhibited a similar expression pattern in crowded and solitary conditions. For example, Ci, Cdc42 and Notch cluster as one group and showed higher expression values at all stages. uba1, Dll, Asc, Scr and Wnt2 were another group, Foxo, Srf, Hh, Ap, Hth, and Tsh were another group, and Fng, Nub, Brk, Antp, Ubx and Dpp formed an additional group. These three groups showed similar expression patterns: the expression in the first and second instars is almost the same, showing slightly lower expression levels; the third to adult stages exhibited higher expression values; and the first group showed higher expression levels relative to the rest of the groups. Vg, Rho, En, Ser, Wg, and Wnt1 and Al, Omb, Spalt, Ptc, Wnt11, and Wnt16 form the remaining two groups. They both showed a similar trend of expression, displaying lower expression values at all stages. Additionally, Fl exhibited a differentially expressed pattern compared to other genes, becoming a lone group, where the first to third instars showed lower expression values and the fourth instar and adult stages exhibited higher expression. Moreover, comparing the gene expression of apterae and alatae at the same stage of aphids, we found that most of the genes, i.e., Ci, Cdc42, uba1, Dll, Asc, Scr, Wnt2, Foxo, Ap, Hth, Tsh, Fng, Brk, Antp, Ubx, Dpp, En, Ser, Wg, Wnt1, Al, Spalt and Ptc, exhibited higher expression in apterae relative to alatae, while Fl exhibited higher expression levels in alatae than apterae. In addition, the rest genes, such as Notch, Srf, Hh, Nub, Vg, Rho, Omb, Wnt11 and Wnt16, showed different expression values in terms of alatae and apterae.
Heat map of wing-associated gene expression profiling among offspring in crowded and solitary conditions by RNA-seq. Genes were clustered via MEV using average FPKM expression values for each of the three replicates. Expression values are relative to one another within each gene, with red representing the highest and the green the lowest values.
Discussion
Environmental factors can affect the wing polyphenism, crowded conditions were sufficient for high production of winged offspring by a group of aphids, while aphids reared under solitary treatment conditions never produced winged aphids or produced fewer winged offspring, indicating that crowded or solitary conditions result in drastic changes for aphids and are conditions that require gene expression responses. This result was consistent with A. pisum study17. Thus, although we treated the aphids equally, individuals probably did not receive the same level of wing-inducing signal during the 16 h of crowded or solitary stimulation. Despite equal treatment, the differential cue reception may be part of a stochastic polyphenism strategy, in which, once induced, aphids produce a stochastic output of winged and wingless morphs. This type of bet-hedging strategy is thought to be used in cases where environmental predictability is variable23, and it is possible that the relative gene expression changes in response to a biological property of the polyphenism.
Biogenic amines are known to play roles in regulating aspects of locust phase polyphenism24. Additionally, studies of insects, such as crickets, planthoppers and A. pisum, have revealed that alternative wing morphs develop in response to various environmental cues2,8,25,26,27,28,29, and responses to these cues may be mediated by hormones8,9,27,30,31. Therefore, we investigated whether biogenic amines and hormones were potentially involved in controlling R. padi wing polyphenism. In our study, biogenic amines and hormones showed a similar expression pattern in crowded and solitary conditions. For the biogenic amines, serotonin and octopamine showed varied expression levels in terms of different developmental stages, while dopamine showed lower expression levels for all instars. Biogenic amines associated with dopamine, serotonin and octopamine showed varied expression levels in different stages of aphids, indicating a possible functional role in wing phenotype determination through changes in the timing or expression level of transporters or receptors in different development stages in response to stressful environmental cues. Biogenic amines are small molecules synthesized from amino acids within neurons of the central nervous system. They are an integral part of the neuroendocrine system32. Biogenic amine signaling is of particular interest with respect to polyphenisms, as it plays a causal role in the phase polyphenism induced by crowding in locusts and other insects in response to stressful environmental cues33,34,35,36. Our results are consistent with previous studies, suggesting that transmission of environmental signals via biogenic amines is a possible factor in density-induced polyphenism and may be a common mechanism generally underlying aphid polyphenism.
Additionally, hormones related to juvenile hormone, ecdysone and insulin exhibited a varied trend of expression patterns at different stages of aphids, indicating that they may play roles in regulating aspects of wing polyphenism through changes in their timing or the expression of hormone enzymes, proteins, transporters or receptors in different development stages in response to environmental stimuli. Hormones are strong candidates for mediating the maternal response to crowding; hormone signals can traverse the hemolymph and affect the organism at the systemic level, and the polyphenism is marked by systemic differences between the morphs. Our results, combined with Braendle et al.’s study9, suggest that hormones possibly have an effect on aphid wing polyphenism. Combined with biogenic amines and hormone studies, we suggested a possible mechanism by which polyphenic aphids may have built upon density stress responses to influence alternative wing development.
To identify genes that could be involved in morphological divergence in aphid wing development, we examined the transcriptomes of all instars at five sequential developmental stages, and we found thirty-three differentially expressed genes. To further study, we found that these genes play well-known roles in anterior-posterior axis determination, dorsal-ventral axis formation, segmentation and wing hinge growth in insect wing development37,38,39,40,41,42,43,44,45,46,47 and likely perform wing-associated functions in aphids, potentially even playing roles in wing development. Therefore, we examined the expression levels of these wing-associated genes via hierarchical clustering at all developmental stages, and we obtained a preliminary heat map showing the timing and level of their expression. Most genes showed a similar trend in expression patterns. We observed a tendency of several genes, i.e., uba1, Dll, Asc, Scr, Wnt2, Foxo, Srf, Hh, Ap, Hth, Tsh, Fng, Nub, Brk, Antp, Ubx and Dpp, to be expressed at lower levels in the first and second instars but higher expression values during the third instar to adult stages. Another group of genes, including Vg, Rho, En, Ser, Wg, Wnt1, Al, Omb, Spalt, Ptc, Wnt11 and Wnt16, showed lower expression values during all stages of aphids. A small number of genes, such as Ci, Cdc42 and Notch, exhibited higher expression levels for all instars. Additionally, Fl showed a different expression pattern compared to the other genes, with lower expression values during the first to third instars and higher expression levels during the fourth instar and adult stages. According to the above gene expression results, we found that eighteen genes, i.e., Wnt2, Fng, Uba1, Hh, Foxo, Dpp, Brk, Ap, Dll, Hth, Tsh, Nub, Scr, Antp, Ubx, Asc, Srf and Fl, had varied expression values during different development stages, indicating that they may play roles in regulating aspects of wing polyphenism through changes in their timing or expression in certain development stages of the aphid.
Interestingly, comparing the gene expression patterns of apterae and alatae at the same life stages to further analysis these eighteen wing-associated genes, we found that most genes, such as Wnt2, Fng, Uba1, Foxo, Dpp, Brk, Ap, Dll, Hth, Tsh, Scr, Antp, Ubx and Asc, exhibited higher expression levels in apterae relative to alatae, although Fl exhibited higher expression levels in alatae than apterae. Additionally, a few genes, such as Srf, Hh and Nub, showed different expression values in terms of alatae and apterae. These genes have well-established functions in the wing development of insects, such as Drosophila, Bombyx mori, Myzus persicae, M. crassicauda, Aphis gossypii and A. pisum12,13,14,15,16,17,37,38,39,40,41,42,43,44,45,46,47,48. Together with previous studies, we discuss the functions of genes in regulating aspects of wing polyphenism in R. padi below.
To further study, we found that several genes were participated in signaling pathways, such as Wnt, Notch, Hh, Ecdysone, Insulin, and Dpp signaling pathways. These signaling pathways, which have previously identified functions in insect wing development40,41,49,50,51,52,53,54,55,56, are likely to play roles in aphid wing development. Several of the genes identified in this study are signal regulators, e.g., Wnt2 is a member of the WNT gene family, which consists of structurally related genes that encode secreted signaling proteins involved in the Wnt signaling pathway, and studies of silkworms have shown that Wnt2 plays a role in the regulation of wing development53,54. Fng is a boundary-specific signaling molecule that encodes secreted proteins involved in the Notch signaling pathway that are required for wing and margin formation, and Fng mediates interactions between dorsal and ventral cells during Drosophila wing development49. Additionally, Uba1, which is indirectly related to the Notch and Wnt signaling pathways, is known to catalyze the first step in ubiquitination, which is crucial for protein degradation55,56. A study in Drosophila proved that the loss of function of Uba1 caused autonomous cell-cycle arrest57, and ubiquitination has been implicated in several signal transduction cascades, including the Notch and Wnt signaling pathways58,59. Thus, Uba1 may be responsible for the temporal-specific activation of signaling pathways involved in wing-morph determination through ubiquitin-mediated control of cell proliferation. Hh is a signaling protein that participates in the Hh signaling pathway. It is specifically expressed in posterior compartment cells and plays a key role in patterning Drosophila imaginal discs40,41. Foxo is a subgroup of the Forkhead family of transcription factors that regulates the transcription of genes regulating diverse cellular processes60 as well as endocrine signaling proteins involved in the insulin signaling pathway. Studies of M. crassicauda proved that Foxo plays a role in wing-morph determination15. In addition, Dpp and Brk are signaling regulators of the Dpp signaling pathway. Studies of Drosophila have shown that the growth and patterning of the wing is controlled in part by the long-range organizing activities of Dpp50,51,52. Dpp is synthesized by cells that line the anterior side of the anterior or posterior compartment border of the wing imaginal disc. Dpp and Brk are expressed in different patterns and are thought to generate a concentration gradient that separates the anterior and posterior compartments to control wing development61,62. Brk encodes a protein that, as a Dpp target, negatively regulates Dpp signaling, and Brk expression is repressed by Dpp. In Drosophila, Brk can also repress the targets of a vertebrate homologue of Dpp, indicating that Brk underscores the importance of its negative role in regulating Dpp activity63. Thus, Brk may be involved in aphid wing determination.
Other identified genes are involved in major wing patterning events, e.g., dorsal-ventral (D-V) patterning genes, such as Ap and Dll38,39,42. Previous studies of Drosophila38,64,65 have shown that Ap is a member of the LIM family of developmental regulatory genes required for the normal development of the wing and haltere imaginal discs. Combined with research on pea aphids14, Dll is one of the wing-patterning genes that regulate wing development. Our results are consistent with previous studies, suggesting that Ap and Dll may play roles in differentiating the morphs. The wing hinge development genes, such as Hth, Tsh, and Nub14, were also identified; in Drosophila, Hth, Tsh and Nub function in the development of the wing hinge43,45. In addition, research on pea aphids14 has shown that Hth, Tsh and Nub are regulated genes participating in wing development in aphids. Additionally, Hox genes, i.e., Scr, Antp, and Ubx, also participate in wing patterning events14,37,44. In Drosophila, wings and halteres are the dorsal appendages of the second and third thoracic segments, respectively. In the third thoracic segment, wing development is suppressed by the homeotic selector gene Ubx to mediate haltere development66. Loss of Ubx function from developing haltere discs induces haltere-to-wing transformation, whereas ectopic expression of Ubx in developing wing discs leads to wing-to-haltere transformation66,67,68. Additionally, research on pea aphids14 suggested that Scr and Antp are regulated genes that participate in wing development, but their functions are not clear. Studies in Drosophila revealed that Scr, Antp and Ubx are homeotic genes in the thorax37,39; thus, they likely play roles in wing development.
The remaining identified genes, i.e., Asc, Srf and Fl, produce factors that regulate morphological features that likely play roles in regulating wing polyphenism. Asc encodes several homologous polypeptides that contain the BHLH (basic helix-loop-helix) domain, representing a subset of a complex gene family69,70,71. Four homologous genes were identified as Asc in silkworms48, expressing significantly higher levels in wing buds and indicating that Asc plays an important role in the formation of wing development. Srf is a member of the MADS-box family of transcription factors involved in orchestrating disparate programs of gene expression linked to muscle differentiation and cellular growth72,73,74. Srf encodes a MADS-box containing a transcriptional regulator, which is expressed in the intervein tissue of wing imaginal discs in Drosophila75, demonstrating that Srf plays a dual role during wing differentiation and acts in a dose-dependent manner to suppress the formation of wing veins. Srf is autonomously required in cells to promote the development of intervein cells. Previous studies on Drosophila, planthoppers and pea aphids13,76,77,78,79,80 suggest that Fl is a myosin rod-binding component of indirect flight muscles and is an essential part of the flight muscle contractile mechanism for thick filament assembly and sarcomere stability. In our study, Fl showed higher expression levels in wing morphs of fourth instar and adult stages, indicating that Fl likely plays an important role in the morphological divergence of the wing morphs.
Overall, through the timing and level of gene expression at all the developmental stages of aphids, we found that most genes showed a similar trend in the expression patterns, except for several genes that showed higher or lower expression values during all stages. For example, uba1, Dll, Asc, Scr, Wnt2, Foxo, Srf, Hh, Ap, Hth, Tsh, Fng, Nub, Brk, Antp, Ubx and Dpp were expressed at lower levels in the first and second instars but exhibited higher expression values in the third instar to adult stages. These results suggest that these genes may play roles in regulating aspects of wing polyphenism through changes in their timing or level of expression in certain stages of aphids. Moreover, we found that the strongest differences in gene expression in winged morphs occurred in the second instar and third instar nymphs, i.e., genes were expressed at lower levels in the second instar and exhibited higher expression values in the third instar, implying that the third instar is a critical stage for visibility of alternative morphs.
Materials and Methods
Insect rearing and sample preparation
A single apterous viviparous parthenogenetic female R. padi was collected from a laboratory at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China, in March of 2016. Samples colonies (voucher number 24644) generated from the above individual had been established in an incubator under long-day conditions (16 hours light: 8 hours dark, 22 °C) and a relative humidity of 65% on wheat seedlings for one year before being sacrificed in subsequent experiments. Samples from the same clonal colony preserved in 75% ethanol were maintained for slide-mounted voucher specimens for morphological identification. Species identification was performed by Ge-Xia Qiao and Li-Yun Jiang based on exterior morphological features of slide-mounted specimens. Prior to the start of all experiments, parthenogenetic females were maintained at a low density (one per plant) on wheat seedlings for ten generations to eliminate cross-generational effects on offspring morph determination81.
Environmental factors affecting the wing polyphenism during parthenogenetic generations are widely known to be influenced by density (contact stimuli), nutritional conditions (plant quality), temperature and photoperiod9,81,82,83. In this study, we used crowded and solitary conditions as wing-inducing stimulation. In the eleventh generation, the parthenogenetic females used for the following experiments were adults. The crowd-induced winged aphids were generated by placing twenty parthenogenetic females together in a square petri dish (length: 2 cm, width: 2 cm) without food. An equal number of parthenogenetic females were placed individually in a square petri dishes without food (one per square petri dish) for solitary treatment. Parthenogenetic females were subjected to crowded or solitary treatments for 16 h. Then, they were transferred to wheat seedlings (one per plant), allowed to produce nymphs for 24 h, and monitored for molting every day to collect all the developmental stages of the aphids.
The first instar nymphs were directly collected from the crowded and solitary mother parthenogenetic females at 24 h, i.e., the “CR24644-1” and “SOL24644-1” samples. Similarly, the nymphs produced in the first 24 h were allowed to develop until the first molt, and the second instar nymphs were harvested, i.e., “CR24644-2” and “SOL24644-2” samples. Starting with the third instar nymphs, small wing buds were identified. For the fourth instar and adult stages, wing buds were outwardly visible. We collected the winged third instar and fourth instar and adult stage aphids from parthenogenetic females that experienced crowded conditions and produced more than 90% winged offspring, i.e., the “CRAL24644-3”, “CRAL24644-4” and “CRAL24644-5” samples. At the same time, a small amount of the third instar and fourth instar and adult stage aphids without visible wing buds were collected, i.e., the “CRAP24644-3”, “CRAP24644-4” and “CRAP24644-5” samples. We then collected the wingless third instar and fourth instar and adult stage aphids from parthenogenetic females reared under solitary conditions that produced more than 90% wingless offspring, i.e., the “SOLAP24644-3”, “SOLAP24644-4” and “SOLAP24644-5” samples. A small number of winged third instar and fourth instar and adult stage aphids were collected in the same manner, i.e., the “SOLAL24644-3”, “SOLAL24644-4” and “SOLAL24644-5” samples. All aphids were harvested during the same time of day to avoid any effects of photoperiod on the collection (Fig. 3).
Aphid rearing and sample collection. As detailed in the methods, sixteen sets of aphids were used in this study. First, aphids were subjected to a solitary or crowded environment for 16 h respectively. Then, after 16 h of solitary or crowded environments, aphids were individually returned to wheat seedlings to feed and monitored for molting every day to collect all the developmental stages of aphids.
In total, we collected individuals for all developmental stages of the offspring for crowded and solitary treatments (the first instar with 25 individuals, second instar with 12 individuals, third instar with eight individuals, fourth instar with six individuals and adult with five individuals), with three replicates for a total of 48 samples (Table S7). All samples (adult stages aphids were dissected to remove and discard ovaries with their developing embryos) were immediately frozen in liquid nitrogen and stored permanently at −80 °C.
RNA extraction and library construction
Total RNA was isolated from pooled whole-body tissues of the first, second, third, fourth and adult stages aphids using an RNeasy mini Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions. Then, RNA was assessed for purity and integrity using an Agilent 2100 Bioanalyzer and an ABI StepOnePlus Real-Time PCR System (Agilent Technologies). cDNA libraries were sequenced in paired-end modes on an Illumina HiSeq 4000 system. A total of 48 libraries were constructed with three biological replicates per sample, and the reference gene library was constructed by mixing equal quantities of total RNA from all samples.
De novo assembly and read mapping
The raw reads were obtained after sequencing and subsequently filtered to obtain clean reads. Then, clean reads were used in de novo assembly and read-mapping in comparison to the reference genes. The RNA-Seq data were de novo assembled using the SOAP program84, and sequences of the unigenes were produced by Trinity v2.0.685. After that, unigenes from the reference gene library were further spliced and assembled to obtain non-redundant unigenes by Tgicl v2.0.686. An additional set of 48 libraries was also filtered in the same manner using the SOAP program84 to obtain clean reads, then Bowtie221 was used to map clean reads to the reference gene to obtain mapped reads.
Functional annotation and gene expression
Functions of the unigenes were annotated by Blast v2.2.2387 with a cutoff E-value of 1E−5 to NCBI databases, including the Nr, Nt, Swissprot, Clusters of Orthologous Groups of proteins (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases88. Additionally, based on the Nr annotation, Gene ontology (GO) classification was obtained by WEGO89 via GO IDs annotated by Blast2GO v2.5.090. An alignment package, SOAP aligner (Version 2.20)84, was used to map reads back to the reference genes. Additionally, we mapped clean reads to the reference genes using Bowtie221 in the first, second, third, fourth and adult stage aphids, and the relative expression levels of all the matched unigenes were normalized by transforming the clean data to FPKM91,92. Comparisons were made between all developmental stages of the offspring, and differentially expressed genes were screened by the NOISeq method93. Moreover, a fold change (FC) ≥ 2 and a probability ≥0.8 were used to filter significantly differentially expressed genes94.
Data Availability
All data used in this manuscript are present in the manuscript and its supporting information. Additionally, the accession numbers of raw data will be available only after acceptance of the manuscript for publication.
References
West-Eberhard, M. J. Developmental plasticity and evolution. (Oxford University Press, 2003).
Nijhout, H. F. Control mechanisms of polyphonic development in insects. BioScience 49, 181–192 (1999).
Nijhout, H. F. Development and evolution of adaptive polyphenisms. Evol. Dev. 5, 9–18 (2003).
Moczek, A. P. Phenotypic plasticity and diversity in insects. Phil. Trans. R. Soc. B 365, 593–603 (2010).
Moczek, A. P. et al. The role of developmental plasticity in evolutionary innovation. P. Roy. Soc. B 278, 2705–2713 (2011).
Snell-Rood, E. C., van Dyken, J. D., Cruickshank, T., Wade, M. J. & Moczek, A. P. Toward a population genetic framework of developmental evolution: the costs, limits, and consequences of phenotypic plasticity. BioEssays 32, 71–81 (2010).
Remaudière, G. & Remaudière, M. Catalogue des Aphididae du monde: Homoptera Aphidoidea. (Institut National de la Recherche Agronomique, 1997).
Simpson, S. J., Sword, G. A. & Lo, N. Polyphenism in insects. Curr. Biol. 21, R738–R749 (2011).
Braendle, C., Davis, G. K., Brisson, J. A. & Stern, D. L. Wing dimorphism in aphids. Heredity 97, 192–199 (2006).
Ishikawa, A., Hongo, S. & Miura, T. Morphological and histological examination of polyphenic wing formation in the pea aphid Acyrthosiphon pisum (Hemiptera, Hexapoda). Zoomorphology 127, 121–133 (2008).
Shi, S. L., Liu, X. X., Zhang, Q. W. & Zhao, Z. W. Morph-specific differences in metabolism related to flight in the wing-dimorphic Aphis gossypii. Insect Sci. 17, 527–534 (2010).
Ghanim, M., Dombrovsky, A., Raccah, B. & Sherman, A. A microarray approach identifies ANT, OS-D and takeout-like genes as differentially regulated in alate and apterous morphs of the green peach aphid Myzus persicae (Sulzer). Insect Biochem. Mol. Biol. 36, 857–868 (2006).
Brisson, J. A., Davis, G. K. & Stern, D. L. Common genome-wide patterns of transcript accumulation underlying the wing polyphenism and polymorphism in the pea aphid (Acyrthosiphon pisum). Evol. Dev. 9, 338–346 (2007).
Brisson, J. A., Ishikawa, A. & Miura, T. Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Mol. Biol. 19, 63–73 (2010).
Ishikawa, A. et al. Screening of upregulated genes induced by high density in the vetch aphid Megoura crassicauda. J. Exp. Zool. 317, 194–203 (2012).
Yang, X. et al. Gene expression profiling in winged and wingless cotton aphids, Aphis gossypii (Hemiptera: Aphididae). Int. J. Biol. Sci. 10, 257–267 (2014).
Vellichirammal, N. N., Madayiputhiya, N. & Brisson, J. A. The genomewide transcriptional response underlying the pea aphid wing polyphenism. Mol. Ecol. 25, 4146–4160 (2016).
Jiang, L. Y., Qiao, G. X., Zhang, G. X. & Zhong, T. S. Fauna of Agricultural and Forestry Aphids in Northeast China (Science Press, 2011).
Richards, W. R. A synopsis of the genus Rhopalosiphum in Canada. Can. Entomol. 92, 1–51 (1960).
Torikura, H. Revisional notes on Japanese Rhopalosiphum, with keys to species based on the morphs on the primary host. Entomol. Soc. Jpn. 59, 257–273 (1991).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Saeed, A. I. et al. TM4: a free, open source system for microarray data management and analysis. BioTechniques 34, 374–378 (2003).
Walker, T. J. Stochastic polyphenism: coping with uncertainty. Fla. Entomol. 69, 46–62 (1986).
Wang, X. & Kang, L. Molecular mechanisms of phase change in locusts. Annu. Rev. Entomol. 59, 225–244 (2014).
Zera, A. J. Wing polymorphism in Gryllus (Orthoptera: Gryllidae): proximate endocrine, energetic and biochemical mechanisms underlying morph specialization for flight vs. reproduction. (Science Press, 2009).
Zera, A. J. & Denno, R. F. Physiology and ecology of dispersal polymorphism in insects. Annu. Rev. Entomol. 42, 207–230 (1997).
Denno, R. F., Douglas, L. W. & Jacobs, D. Crowding and host plant nutrition: environmental determinants of wing-form in Prokelisia marginata. Ecology 66, 1588–1596 (1985).
Braendle, C., Friebe, I., Caillaud, M. C. & Stern, D. L. Genetic variation for an aphid wing polyphenism is genetically linked to a naturally occurring wing polymorphism. P. Roy. Soc. B 272, 657–664 (2005).
Roff, D. A. & Fairbairn, D. J. The evolution and genetics of migration in insects. BioScience 57, 155–164 (2007).
Lees, A. D. Action of juvenile hormone mimics on the regulation of larval-adult and alary polymorphism in aphids. Nature 267, 46–48 (1977).
Schwartzberg, E. G., Kunert, G., Westerlund, S. A., Hoffmann, K. A. & Weisser, W. W. Juvenile hormone titres and winged offspring production do not correlate in the pea aphid, Acyrthosiphon pisum. J. Insect Physiol. 54, 1332–1336 (2008).
Roeder, T. Biogenic-amines and their receptors in insects. Comp. Biochem. Physiol. C 107, 1–12 (1994).
Iba, M., Nagao, T. & Urano, A. Effects of population density on growth, behavior and levels of biogenic amines in the cricket, Gryllus bimaculatus. Zool. Sci. 12, 695–702 (1995).
Hirashima, A., Sukhanova, M. & Rauschenbach, I. Biogenic amines in Drosophila virilis under stress conditions. Biosci. Biotech. Bioch. 64, 2625–2630 (2000).
Chen, Y. L., Hung, Y. S. & Yang, E. C. Biogenic amine levels change in the brains of stressed honeybees. Arch. Insect Biochem. Physiol. 68, 241–250 (2008).
Wada-Katsumata, A., Yamaoka, R. & Aonuma, H. Social interactions influence dopamine and octopamine homeostasis in the brain of the ant Formica japonica. J. Exp. Biol. 214, 1707–1713 (2011).
Struhl, G. Genes controlling segmental specification in the Drosophila thorax. PNAS 79, 7380–7384 (1982).
Cohen, B., McGuffin, M. E., Pfeifle, C., Segal, D. & Cohen, S. M. Apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Gene Dev. 6, 715–729 (1992).
Campbell, G., Weaver, T. & Tomlinson, A. Axis specification in the developing Drosophila appendage: the role of wingless, decapentaplegic, and the homeobox gene aristaless. Cell 74, 1113–1123 (1993).
Basler, K. & Struhl, G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 368, 208–214 (1994).
Tabata, T. & Kornbert, T. B. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 76, 89–102 (1994).
Kim, J., Irvine, K. D. & Carroll, S. B. Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell 82, 795–802 (1995).
Sturtevant, M. A. & Bier, E. Analysis of the genetic hierarchy guiding wing vein development in Drosophila. Development 121, 785–801 (1995).
Weatherbee, S. D., Halder, G., Kim, J., Hudson, A. & Carroll, S. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Gene Dev. 12, 1474–1482 (1998).
Butler, M. J. et al. Discovery of genes with highly restricted expression patterns in the Drosophila wing disc using DNA oligonucleotide microarrays. Development 130, 659–670 (2003).
Ren, N., Zhu, C., Lee, H. & Adler, P. N. Gene expression during Drosophila wing morphogenesis and differentiation. Genetics 171, 625–638 (2005).
Kiger, J. A. et al. Tissue remodeling during maturation of the Drosophila wing. Dev. Biol. 301, 178–191 (2007).
Tong, X. L. Study on the wing pattern genes in the silkworm, Bombyx mor. (Southwest University, 2008).
Irvine, K. & Wieschaus, E. Fringe, a boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell 79, 595–606 (1994).
Capdevila, J. & Cuerrero, I. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO. J. 13, 4459–4468 (1994).
Nellen, D., Burke, R., Struhl, G. & Basler, K. Direct and long-range action of a DPP morphogen gradient. Cell 85, 357–368 (1996).
Lecuit, T. et al. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature 381, 387–393 (1996).
Cadigan, K. M. & Nusse, R. Wnt signaling: a common theme in animal development. Gene Dev. 11, 3286–3305 (1997).
Miller, J. R. The Wnts. Genome Biol. 3, reviews3001 (2001).
Pickart, C. M. Mechanism underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 (2001).
Kulkarni, M. & Smith, H. E. E1 ubiquitin-activating enzyme Uba-1 playsmultiple roles throughout C. elegans development. PLoS Genet. 4, e1000131 (2008).
Lee, T. V. et al. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development 135, 43–52 (2008).
Lai, E. C., Roegiers, F., Qin, X., Jan, Y. N. & Rubin, G. M. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development 132, 2319–2332 (2005).
Tauriello, D. V. F. & Maurice, M. M. The various roles of ubiquitin in Wnt pathway regulation. Cell Cycle 9, 3700–3709 (2010).
Carter, M. E. & Brunet, A. FOXO transcription factors. Curr. Biol. 17, R113–R114 (2007).
Masucci, J. D., Miltenberger, R. J. & Hoffmann, F. M. Pattern-specific expression of the Drosophila decapentaplegic gene in imaginal disks is regulated by 3′ cis-regulatory elements. Gene Dev. 4, 2011–2023 (1990).
Kaphingst, K. & Kunes, S. Pattern formation in the visual centers of the Drosophila brain: wingless acts via decapentaplegic to specify the dorsoventral axis. Cell 78, 437–448 (1994).
Minami, M., Kinoshita, N., Kamoshida, Y., Tanimoto, H. & Tabata, T. Brinker is a target of Dpp in Drosophila that negatively regulates Dpp-dependent genes. Nature 398, 242–246 (1999).
Bourgouin, C., Lundgren, S. E. & Thomas, J. B. Apterous is a Drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron 9, 549–561 (1992).
Lundgren, S. E., Callahan, C. A., Thor, S. & Thomas, J. B. Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development 121, 1769–1773 (1995).
Lewis, E. B. A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 (1978).
Cabrera, C. V., Botas, J. & Garcia-Bellido, A. Distribution of Ultrabithorax proteins in mutants of Drosophila bithorax complex and its transregulatory genes. Nature 318, 569–571 (1985).
White, R. A. H. & Akam, M. Contrabithorax mutations cause inappropriate expression of Ultrabithorax in Drosophila. Nature 318, 567–569 (1985).
Alonso, M. C. & Cabrera, C. V. The achaete-scute gene complex of Drosophila melanogaster comprises four homologous genes. EMBO. J. 7, 2585–2591 (1988).
Ghysen, A. & Dambly-Chaudière, C. From DNA to form: the achaete-scute complex. Gene Dev. 2, 495–501 (1988).
González, F., Romani, S., Cubas, P., Modolell, J. & Campuzano, S. Molecular analysis of the asense gene, a member of the achaete-scute complex of Drosophila melanogaster, and its novel role in optic lobe development. EMBO. J. 8, 3553–3562 (1989).
Shore, P. & Sharrocks, A. D. The MADS-box family of transcription factors. FEBS. J. 229, 1–13 (1995).
Fristrom, D. et al. Blistered: a gene required for vein/intervein formation in wings of Drosophila. Development 120, 2661–2671 (1994).
Miano, J. M. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 35, 577–593 (2003).
Montagne, J. et al. The Drosophila Serum Response Factor gene is required for the formation of intervein tissue of the wing and is allelic to blistered. Development 122, 2589–2597 (1996).
Vigoreaux, J. O., Saide, J. D., Valgeirsdottir, K. & Pardue, M. L. Flightin, a novel myofibrillar protein of Drosophila stretch-activated muscles. J. Cell Biol. 121, 587–598 (1993).
Vigoreaux, J. O., Hernandez, C., Moore, J., Ayer., G. & Maughan, D. A genetic deficiency that spans the flightin gene of Drosophila melanogaster affects the ultrastructure and function of the flight muscles. J. Exp. Biol. 201, 2033–2044 (1998).
Reedy, M. C., Bullard, B. & Vigoreaux, J. O. Flightin is essential for thick filament assembly and sarcomere stability in Drosophila flight muscles. J. Cell Biol. 151, 1483–1499 (2000).
Ayer, G. & Vigoreaux, J. O. Flightin is a myosin rod binding protein. Cell Biochem. Biophys. 38, 41–54 (2003).
Xue, J. et al. Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS ONE 5, e14233 (2010).
Sutherland, O. R. W. The role of crowding in the production of winged forms by two strains of the pea aphid, Acyrthosiphon pisum. J. Insect Physiol. 15, 1385–1410 (1969).
Lees, A. D. The control of polymorphism in aphids. Adv. Insect Physiol. 3, 207–277 (1966).
Müller, C. B., Williams, I. S. & Hardie, J. The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecol. Entomol. 26, 330–340 (2001).
Li, R. Q. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Manfred, G. G. et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652 (2011).
Pertea, G. et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652 (2003).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y. & Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280 (2004).
Ye, J. et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34, W293–W297 (2006).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Morozova, O., Hirst, M. & Marra, M. A. Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genom. Hum. G. 10, 135–151 (2009).
Oshlack, A., Robinson, M. D. & Young, M. D. From RNA-seq reads to differential expression results. Genome Biol. 11, 220 (2010).
Tarazona, S., García-Alcalde, F., Dopazo, J., Ferrer, A. & Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 21, 2213–2223 (2011).
Abdi, H. The bonferroni and sidak corrections for multiple comparisons. (Encyclopedia of Measurement and Statistics, 2007).
Acknowledgements
We would like to thank Ms. Ju-Lian Chen for providing the live aphids reared in this research, thanks are also given to Ms. Fen-Di Yang for making the slide-mounted specimens. This study was supported by the National Natural Science Foundation of China (Nos 31620103916 and 31430078), and National Key R&D Program of China (Grant No. 2016YFE0203100).
Author information
Authors and Affiliations
Contributions
G.Q., J.C., R.Z. and L.J. designed the research; R.Z. performed the research; and L.J. and G.Q. identified the voucher specimens. R.Z. and J.C. provided and analyzed the data, R.Z. wrote the paper, and all authors examined and approved the final version of the manuscript for publication.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, RJ., Chen, J., Jiang, LY. et al. The genes expression difference between winged and wingless bird cherry-oat aphid Rhopalosiphum padi based on transcriptomic data. Sci Rep 9, 4754 (2019). https://doi.org/10.1038/s41598-019-41348-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41348-1
This article is cited by
-
Insights into wing dimorphism in worldwide agricultural pest and host-alternating aphid Aphis gossypii
Journal of Cotton Research (2021)
-
Differences among families in craniofacial shape at early life-stages of Arctic charr (Salvelinus alpinus)
BMC Developmental Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.