Abstract
SOX6 is a HMG-box transcription factor expressed in a wide range of tissues. Recent data show that SOX6 expression is altered in different cancers, in the majority of cases being downregulated. To date, no data are available about SOX6 role in hematological malignancies. Here we demonstrate that SOX6 overexpressing BCR-ABL1+ B-ALL cells are unable to promote leukemia in a mouse model. Starting from this observation, we extended our study to a panel of human leukemic cells carrying genetic lesions distinctive of different types of leukemias and myeloproliferative disorders (the BCR-ABL1 translocation and the JAK2V617F amino acid substitution) to dissect the cellular events induced by SOX6. The inhibition of proliferation is the invariant outcome of SOX6 overexpression but it is achieved via two different cellular responses: terminal differentiation in erythroid-biased cells, irrespectively of their mutation, and apoptosis in megakaryocytic-primed and lymphoid cells. Within this context, cells carrying the highest copy number of the JAK2V617F allele better counteract the SOX6-imposed growth arrest. The interrogation of the GEPIA (Gene Expression Profiling Interactive Analysis) human dataset reveals that SOX6 is downregulated in a cohort of AML patients, uncovering a wide anti-proliferative role of SOX6 in a variety of mutant backgrounds.
Similar content being viewed by others
Introduction
The SOX6 transcription factor, belonging to the Sry-related HMG-box family, is expressed in several tissues during development, where it plays a key role in the transition from proliferating progenitors to functionally mature cells1. In the adult, its role is more elusive.
Data collected in recent years suggest that SOX6 expression is deregulated in different cancers. Often, SOX6 seems to act as tumor suppressor such as in Esophageal Squamous Cell Carcinoma -ESCC-2, Hepatocarcinoma -HCC-3,4, ovarian cancer5, pancreatic cancer6, colorectal cancer7 and its downregulation is frequently associated with a poor prognosis2,4,5. Rescue experiments performed by SOX6 overexpression in these types of cancers often result in a mitigated malignant phenotype2,5,8,9. In contrast, in other cancers, such as some brain tumors10, SOX6 expression is higher compared to normal tissues. The cause of SOX6 deregulation and, ultimately, the alteration of the cellular events downstream to it, remains largely unknown.
In hematopoiesis, SOX6 is required for proper erythroid terminal differentiation11,12. Its knock out in mouse results in compensated anemia13; its conditional ablation in erythroid cells impairs erythropoiesis in both homeostatic and stress condition11. Conversely, SOX6 overexpression induces a strong hemoglobinization, in both murine and human cellular model systems12,13. Interestingly, we noticed that SOX6 overexpression in cells carrying different genetic lesions, BCR-ABL1+ (e14a2 splicing variant p210BCR-ABL1+) (K562) and JAK2V617F+ (JAK2 c.1849G > T; p.Val617Phe) (HEL), leads to different proliferation kinetics. Indeed, HEL cells keep growing even when the K562 culture is already exhausted12.
The BCR-ABL1 fusion protein is the hallmark of Chronic Myeloid Leukemia (CML)14,15, but it is also a frequent cytogenetic abnormality in precursor B-lymphoblastic leukemia (B-Acute Lymphoblastic Leukemia, B-ALL) found in adults16 and, more rarely, in pediatric patients17. The JAK2V617F mutation is typical of myeloproliferative disorders (MPDs) and Acute Myeloid Leukemia (AML)18.
Here we show that SOX6 overexpression invariably blocks cell proliferation in both BCR-ABL1+ and JAK2V617F+ cells, although the JAK2V617F allele confers a graded resistance to SOX6-induced growth arrest depending on its copy number. The cellular mechanisms through which this is achieved is via terminal differentiation or apoptosis. Indeed, in both genetic backgrounds, SOX6 induces differentiation in erythroid-primed cells whereas it promotes apoptosis in erythro/megakaryocytic and B lymphoid cell types. We propose that the two aforementioned cellular pathways activated by SOX6 may account of its wide anti-proliferative role in different blood cell types. This is further supported by data recently made available19, showing that SOX6 is downregulated in a cohort of AML patients.
Results and Discussion
In recent years, several studies reported a reduced level of SOX6 expression in different cancers. To explore its possible onco-suppressive role in hematological cancers, we first exploited the leukemia model of BCR-ABL1-induced B-ALL, generated in Prof. J. Ghysdael’s laboratory by transducing Bone Marrow cells from Cdkn2a-deficient C57BL/6J mice with a retroviral vector encoding for the e1a2 splicing variant of the p190BCR-ABL1 protein and GFP. Genomic deletions of the Cdkn2a locus are a frequent event in BCR-ABL1-positive ALL and are a prognostic marker for poor long-term outcome20. The presence of GFP allows the tracing of these cells (from here called B-ALL) in recipient mice.
B-ALL were firstly transduced either with viral particles carrying the empty vector (EV) or the SOX6-ΔNGFR (SOX6) cassette. In each experiment, the efficiency of transduction was tested by western blot and flow cytometry (FC) (Fig. S1). In the infected cells, expanded in an in vitro culture, SOX6 overexpression induced a significant block in cell proliferation (Fig. 1a). We then injected 106 cells, infected with a comparable efficiency for both EV and SOX6-ΔNGFR (not shown), into recipient age-matched C57BL/6J mice and monitored the development of leukemia (Fig. 1b,c). As expected, all mice developed leukemia within 9 days from transplantation and were sacrificed at the onset of first symptoms. Injected cells infiltrated in both spleen and bone marrow and represented about 80% of the total cells extracted from these organs, confirming that leukemia was indeed due to B-ALL-GFP+ cells (Fig. 1b,c). Strikingly, ΔNGFR-GFP double positive cells were about 15% in control mice injected with EV-transduced cells but less than 0.5% in mice injected with SOX6-tranduced cells (Fig. 1c). Furthermore, GFP+ ΔNGFR+ cells were absent in cultures expanded ex vivo from the bone marrows of mice injected with SOX6-transduced cells (Fig. 1d). Overall, these data provide a clear evidence that the anti-proliferative pathways activated by SOX6 counteract the leukemic potential of B-ALL cells, even in the presence of the double BCR-ABL1+ and Cdkn2a−/− mutation, a combination that frequently is prognostic of a more aggressive lymphoid leukemia21. Moreover, these results suggest that SOX6 may act as an anti-leukemic factor in a Cdkn2a−/− background. This evidence is particularly interesting since the p14ARF-HDM2-p53 axis is a known mediator of the SOX6 anti-proliferative effect in several tumors8. In order to extend this observation, we selected three human leukemic cell lines carrying the BCR-ABL1 translocation in different lineage background: SUP-B15, a B-cell precursor (e1a2 splicing variant p190BCR-ABL1+ CDKN2A−/− 22); K562 and MEG-01, two erythro/megakaryocytic cell types (e14a2 splicing variant p210BCR-ABL1+ p53−/− 23 and e13a2 splicing variant p210BCR-ABL1+ p53−/− 24, respectively). As shown in Fig. 1e, upper panel, SOX6 overexpression invariably blocks cell proliferation in all the three cell types.
SOX6 blocks leukemia development. (a) Growth kinetics of B-ALL cells transduced either with viral particles carrying the SOX6-IRES-ΔNGFR (SOX6) or with EV-IRES-ΔNGFR (EV) vectors. (b) Analysis of spleens from mice injected with 106 B-ALL transduced cells. Spleens were collected, measured, weighted (histogram bars: mean ± SEM n = 9; *p value < 0.05; **p value < 0.01; ***p value < 0.001) and compared to spleens from PBS-injected control animals (CTRL). (c) The transduced B-ALL engraftment contribution was assessed by tracking GFP+ cells (B-ALL cells) and GFP+ ΔNGFR+ double positive cells (B-ALL cells transduced with either SOX6 or EV) by flow cytometry (histogram bars: mean ± SEM; n = 9; *p value < 0.05; **p value < 0.01; ***p value < 0.001 P). (d) The same analysis was performed on explanted bone marrow cells grown ex vivo for 9 days (D1–D9) (histogram bars: mean ± SEM; n = 3). (e) Transduced cell lines growth curves. Transduced cells were ≥95% in each experiment, Fig. S1 (Graph points: mean ± SEM; n ≥ 3 *p value < 0.05; **p value < 0.01; ***p value < 0.001).
The JAK2V617F mutation confers a graded resistance to the SOX6 anti-proliferative effect
In a previous work, we demonstrated that the erythroleukemic HEL cells behave differently compared to K562 upon SOX6 overexpression. Since HEL cells carry the JAK2V617F amino acid substitution, we hypothesized that the presence of this allele could counteract SOX6-induced growth arrest12.
To reinforce this hypothesis, we took advantage of human erythro/megakaryocytic leukemic cell lines carrying a different copy number of the JAK2V617F allele: HEL (8 copies)25, SET-2 (6 copies and one wt allele)26 and UKE1(2 copies)27.
As shown in Fig. 1e, lower panel, JAK2V617F+ cells indeed exhibited a graded resistance to the anti-proliferative SOX6 action depending on the mutant allele copy number. In fact, HEL cells overexpressing SOX6 grew at the same rate of control cells until day 6, started to decline at day 9, and the culture finally exhausted by day 12. SET-2 cells declined after day 6. Finally, in UKE1 cells, the SOX6 induced arrest in cell proliferation was already detectable at day 3.
SOX6 induces apoptosis in non-erythroid cells
Since different cell types carrying contradistinctive genetic lesions stop growing upon SOX6 overexpression, we further investigated the cause of the proliferation block. We therefore analyzed -in both BCR-ABL1+ and JAK2V617F+ cells- three different parameters: i) the proportion of apoptotic cells (by AnnexinV and 7-AAD staining and subsequent FC analysis); ii) the induced differentiation (by testing the expression of prototypical lineage-specific genes); iii) the cells distribution across the different cell cycle phases (by PI staining and subsequent FC analysis).
As shown in Fig. 2 (upper panel), within the BCR-ABL1+ background, the percentage of SOX6-transduced cells undergoing apoptosis was significantly increased in MEG-01 and -more markedly- in SUP-B15 cells, compared to the EV-transduced control cells. In contrast, SOX6 overexpression did not promote apoptosis in K562 cells.
SOX6 induces apoptosis in non-erythroid biased cell lines. Apoptosis was assessed by staining transduced cells with AnnexinV and 7-AAD. Black: dead cells (AnnV− 7-AAD+); dark grey: late apoptotic (AnnV+ 7-AAD+); light gray: early apoptotic (AnnV+ 7-AAD−); white: live cells (AnnV− 7-AAD−) (histogram bars: mean ± SEM; n ≥ 3). Representative plots for each cell type are shown in Fig. S2.
Instead, the analysis of JAK2V617F+ cells pointed to a graded resistance to apoptosis based on the different copy number of the mutant allele, similarly to what we observed for growth kinetics (Fig. 2, lower panel). In fact, whereas HEL cells, which carry 8 copies, did not show a significant increase of apoptotic cells in response to SOX6 overexpression, SET-2 (6 copies) and UKE1 (2 copies) proportionally increased the level of apoptotic cells.
SOX6 induces differentiation in erythroid cells
SOX6 is involved in the production of mature red blood cells, being a key transcription factor required for the balance between cell cycle withdrawal and erythroid terminal differentiation. Since both K562 and HEL cells are commonly accepted as erythroid cellular model systems, we investigated to what extent the observed arrest in cell proliferation and the absence of apoptosis are linked to the erythroid differentiation program.
Figure 3a (lower panel) shows that HEL cells, where growth arrest occurred more slowly compared to K562, indeed undergo a clear differentiation upon SOX6 overexpression, visible as a reduction in cell size. The analysis of prototypical lineage-specific genes confirmed that upon SOX6 overexpression, HEL and K562 cells increased the expression of erythroid marker genes. In cells with a more promiscuous erythro/megakaryocytic phenotype (MEG-01, SET-2, UKE1), the increase in erythroid genes expression occurred at the expenses of megakaryocytic-affiliated genes (i.e. GPIIb, RUNX1, FLI1, MPL). Surprisingly, in SUP-B15 cells, where the expression of erythroid-specific genes was -and remained- undetectable, SOX6 reduced the expression of the B-lineage master-regulator genes EBF1, PAX5 and of the PAX5 downstream effector BLNK28 (Fig. 3a).
SOX6 induces erythroid genes at the expense of genes affiliated with other lineage programs. (a) Expression level of marker genes upon SOX6 transduction. Fold change variation in different lineage-specific genes (erythroid, megakaryocytic, lymphoid) was assessed by RT-qPCR (n ≥ 3). In the erythroleukemic HEL cells overexpressing SOX6 the enhanced erythroid differentiation was confirmed by cells staining. HBA: Hemoglobin α chain; HBG: Hemoglobin γ chain; ALAS-E: Aminolevulinate Synthase, Erythroid; GPIIb: Glycoprotein IIB; RUNX1: Runt-related transcription factor 1; FLI1: Friend leukemia integration 1 transcription factor; MPL: thrombopoietin receptor; PAX5: Paired Box 5; EBF1: Early B Cell Factor 1; BLNK: B Cell Linker. (b) Cells distribution across the different cell cycle phases analyzed by Propidium Iodide staining (PI). Dark gray: G2/M phase; light gray: S phase; white: G0/G1 phase. Arrows indicate significant reduction of the percentage of cells in S-phase.
Consistently with previous observations, the most erythroid-biased cells, i.e. K562 and HEL (and to a lesser extent SET-2, which express erythroid genes at relatively high levels, not shown) responded to SOX6 by accumulating cells in G0/G1 at the expenses of the S phase (Fig. 3b), mimicking the cell cycle withdrawal steps accompanying late stages of erythroid differentiation29. This redistribution across cell cycle phases was not significant in MEG-01 and was absent in both UKE1 and SUP-B15 (Fig. 3b).
Taken together, these data indicate that within an erythroid context (HEL and K562) SOX6 induces terminal differentiation (Fig. 3) which justifies the block in cellular proliferation (Fig. 1e), in the absence of apoptosis (Fig. 2). On the other hand, in cells that are less biased towards the erythroid fate, SOX6 activates an apoptotic response leading to cell death (Fig. 2).
Collectively, the comparison of cell lines representing different hematological disorders reveals a dual effect elicited by SOX6, i.e. the induction of maturation (HEL and K562 cells) in erythroid cells and/or the induction of apoptosis in promiscuous (MEG-01, UKE1, SET-2) or non-erythroid cell types (B cells).
SOX6 is downregulated in human hematological malignancies
These data collectively point to a broad anti-proliferative effect of SOX6, based on its ability to act via a dual mechanism, in a wide spectrum of cell types.
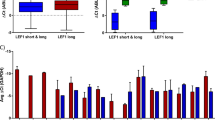
This evidence prompted us to interrogate the GEPIA (Gene Expression Profiling Interactive Analysis) dataset19 to explore SOX6 expression in normal vs. tumoral human hematological samples. Interestingly, SOX6 expression was significantly reduced in a cohort of 173 AML patients (including few BCR-ABL1+ cases) (Fig. 4a). Moreover, the patients overall survival plot revealed a trend suggesting a direct correlation between low levels of SOX6 and a poor prognosis (Fig. 4b).
SOX6 is downregulated in AML patients. (a) GEPIA expression analysis of SOX6 in human AML patients. Gray box: 173 AML patients; white box: 70 healthy donors. (b) Kaplan–Meier curve comparing survival of AML patients with high -compared to low- SOX6 transcript level, based on GEPIA p-value = 0.23 was calculated with log-rank (Mantel–Cox) test.
Taken together, our data uncover an important anti-proliferative effect of SOX6 in a variety of hematological cancer cells, as a result of the induction of pro-differentiation, pro-apoptotic or both pathways. We believe that this is an important aspect in view of understanding the mechanism controlling the onset and/or the progression of cancer characterized by the loss of SOX6, and for the development of new treatment strategies.
Methods
Constructs
The Sox6 murine cDNA (SOX6) was cloned in frame with a flag epitope immediately upstream to the IRES-EmeraldGFP (eGFP) or to the IRES-ΔNGFR cassette of the pHR SIN BX IR/EMW (originally derived from the pHR-SIN-CSGW lentiviral vector)12. The two packaging plasmids psPAX2 and pMD2.GVSVG were used to produce Lentiviral pseudo-particles in 293 T cells (www.lentiweb.com).
Lentiviral production and transduction
Exponentially growing HEK-293T cells were transfected with the calcium-phosphate method with the above vectors plus the packaging plasmids psPAX2 and pMD2.G-VSVG to produce the lentiviral pseudo-particles. 72 h after transfection, the supernatant with recombinant virus particles was collected, filtered and centrifuged at 20,000 g for 8 hours at 4 °C. The viral pellet was re-suspended in 1XPBS and aliquoted at −80 °C. Lentiviruses were titrated on HEK-293T cells by measuring the percentage of eGFP- or of ΔNGFR-positive cells by Flow Cytometry (FC). The transduction of all the cell lines used in this study was performed overnight with a multiplicity of infection (MOI ≥ 30) by adding the viral particles re-suspended in 1X PBS directly to the cellular cultures.
Whole protein extracts
Cells were harvested and centrifuged at 400 g for 5 minutes at room temperature. Pellets were washed twice in ice-cold 1X PBS (Euroclone) and re-suspended in RIPA buffer (20 mM TrisHCl pH 7.4; 150 mM NaCl; 5 mM EDTA; 0,3%TritonX) supplemented with proteases inhibitor cocktail (Roche). Lysis was performed for 30 minutes on ice. After centrifugation at 16000 g for 5 minutes at 4 °C, the supernatant containing protein extracts was retrieved.
Immunoblotting analysis
Total extracts (15–30 μg/lane), resuspended in Laemmli buffer, were resolved by SDS/PAGE in a 10% acrylamide gel and blotted onto Hybond-ECL Nitrocellulose membrane (GE healthcare) under constant voltage at 100 V for 90 min at 4 °C (Biorad Transblot apparatus). Membranes were blocked for 1 hours at room temperature with Milk 5% in 1X TBS-T (Tris Buffered Saline, pH 7.6 and 0,1% Tween20) and incubated with the appropriate primary antibody diluted in Milk 5% 1X TBS-T overnight at 4°C. Membranes were washed three times in 1X TBS-T and incubated with the appropriate HRP-conjugated secondary antibody (in Milk 5% 1X TBS-T) for 1 hours at room temperature. After three washes in 1X TBS-T, antibodies binding was detected by ECL (Millipore). Antibodies used: anti-Flag F7425 (SIGMA) or ab125243 (abcam) and HRP-conjugated horse anti-mouse IgG #7076 (Cell Signaling), to detect the SOX6-FLAG protein; HRP-directly conjugated anti-beta actin #5125 (Cell Signaling) or anti-U2AF #U4758 (Sigma), to detect the loading control protein.
In vivo induction of leukemia in mouse
B-ALL BCR-ABL1+ GFP+ cells were transduced with the SOX6-IRES-ΔNGFR vector or with the corresponding empty vector at a MOI = 50. 24 h later, cells were washed three times with 1X PBS and 106 cells were injected into the tail vein of each C57BL/6 J recipient mouse. An aliquot of these cells was fixed in 4% paraformaldehyde, stained with APC Anti-human CD271 (#345108, BioLegend) for 15 minutes at 4 °C and analyzed for ΔNGFR positivity by flow cytometry. All the experiments and protocols were approved by the Ethical Committee on animal studies (OPBA: Organismo Preposto al Benessere Animale) of the University of Milano-Bicocca and were conducted according to the Italian legislation (approved protocol n°359/2016PR, authorized by the Italian Ministry of Health).
Cell lines
For detailed description, please refer to https://web.expasy.org/cellosaurus/. Briefly: K562: erythroleukemia immortalized myelogenous cell line. MEG-01: human megakaryoblastic leukemic cell line. SUP-B15: Human B cell precursor leukemia. HEL: Human Erythroleukemic cell line. SET-2: human essential thrombocythemic. UKE1: human essential thrombocythemic cells. B-ALL BCR-ABL1+ GFP+: cell line established from Cdkn2a−/− murine bone marrow cells infected with a pMIG retrovirus expressing a p190BCR-ABL1-GFP cassette (a gift from Prof. Ghysdael, Paris Diderot University). K562 and HEL cell lines were grown in RPMI medium (Lonza) supplemented with 10% of FBS (Fetal Bovine Serum) (Sigma); MEG-01 and SET-2 were grown in RPMI medium (Lonza) supplemented with 20% of FBS (Fetal Bovine Serum); UKE1 were grown in IMDM medium (EuroClone) supplemented with 10% FBS (Fetal Bovine Serum) (Sigma) and 10% Horse Serum (Invitrogen), 1uM di HydroCortisone (Sigma); SUP-B15 were grown in RPMI medium (Lonza) supplemented with 10% of FBS (Usa Origin Sigma); B-ALL BCR-ABL1+ GFP+ were grown in RPMI (Lonza) with 15% FBS (Sigma) and 7,5% NaHCO3. The medium of all the cell lines was supplemented with 100 μg/ml penicillin/streptomycin (EuroClone), 4mM L-glutamine (EuroClone). Cells were grown at an optimal concentration of 0,5 × 106 cells/mL, in a humidified 5% CO2 atmosphere at 37 °C.
RNA isolation and Real Time PCR
Total RNA from ≥106 of all different cell lines and experimental condition were purified with TRIzol Reagent (Applied Biosystem), treated with RQ1 DNase (Promega) for 30 minutes at 37 °C and retrotranscribed (High Capacity cDNA Reverse Transcription Kit, Applied Biosystem). Negative control reactions (without Reverse Transcriptase) gave no PCR amplification. Real time analysis was performed using ABI Prism 7500, (Applied Biosystems). Primers were designed to amplify 150 to 300 bp amplicons, spanning an exon-exon junction when possible, on the basis of sequences from the Ensembl database (http://www.ensembl.org). Samples from each experiment were analyzed in triplicate. Specific PCR product accumulation was monitored by using SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad) fluorescent dye in 12-μl reaction volume. Dissociation curves confirmed the homogeneity of PCR products. All primers are available upon request.
Apoptosis Assay
Apoptosis assays were carried out by using AnnexinV (AnnV) and 7-Amino-Actinomycin D (7-AAD) (BD Biosciences). 2,5 × 105 cells were washed twice with cold 1X PBS (Euroclone) and then resuspend in 1X Annexin Binding Buffer (10 mM Hepes pH 7.4), 140 mM NaCl, 2,5 mM CaCl2) with 5 μl of PE Annexin V (BD Biosciences) or APC Annexin V (Biolegend) and 5 μl 7-AAD in a final volume of 100 μL. Cells were gently vortexed and incubated for 15 minutes at RT (25 °C) in the dark. Finally, samples were diluited with 400 μl of 1X Annexin Binding Buffer and analyzed by flow cytometry within 1 hour (Becton-Dickinson FACS Calibur or Beckman-Coulter CytoFLEX S). Data were analyzed with the Summit Software 4.3 or CytExpert Software.
Propidium Iodide (PI) Staininig for cell cycle analysis
Cells, washed twice with cold 1X PBS (Euroclone), were resuspend in 1% paraformaldehyde for 45 minutes. After that, the same volume 1X PBS with Triton 0,2% (Sigma) was added for 15′ minutes. Cells were then centrifuged and resupended in 250 μl of PBS with 0.1 mg/ml PI (Sigma) and 100 μg/mL RNAse (Sigma), vortexed for 5 seconds and incubated at room temperature for at least 1 hour in the dark. Samples were analyzed by flow cytometry within 1 hour (Becton-Dickinson FACS Calibur or Beckman-Coulter CytoFLEX S). Data were analyzed with the Summit Software 4.3 or CytExpert Software.
Cells staining
1,5 × 105 cells were resuspended in 1X PBS with 0,1% BSA and cytospun at 300 rpm for 2 minutes at room temperature. Cells were then fixed with 10% cold Methanol and stained by using the Ematoxylin-Eosin staining kit Diff-Quick dyes (Medium Diagnostics) according to the manufacturer’s protocol.
Statistics
All continuous variables have been tested for normality by using the Shapiro-Wilk test. Experiments on cell lines were analyzed by using a test for paired data while for the in vivo experiments data were analyzed through tests for unpaired data. All tests performed were two-tailed. Statistical analysis was performed by using GraphPad Prism, version 6.0. Data are expressed as mean ± SEM of n ≥ 3 replicates.
Ethical Statement
All the experiments in mice were conducted according to the Italian legislation (protocol n°359/2016PR, authorized by the Italian Ministry of Health).
Data Availability
No datasets were generated in the current study. Figure 4 is based on GEPIA data (Gene Expression Profiling Interactive Analysis, -http://gepia.cancer-pku.cn/-, ref.19).
References
Hagiwara, N. Sox6, jack of all trades: a versatile regulatory protein in vertebrate development. Dev Dyn 240, 1311–1321, https://doi.org/10.1002/dvdy.22639 (2011).
Qin, Y. R. et al. Characterization of tumor-suppressive function of SOX6 in human esophageal squamous cell carcinoma. Clin Cancer Res 17, 46–55, https://doi.org/10.1158/1078-0432.CCR-10-1155 (2011).
Xie, Q. et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer 118, 2431–2442, https://doi.org/10.1002/cncr.26566 (2012).
Guo, X., Yang, M., Gu, H., Zhao, J. & Zou, L. Decreased expression of SOX6 confers a poor prognosis in hepatocellular carcinoma. Cancer Epidemiol 37, 732–736, https://doi.org/10.1016/j.canep.2013.05.002 (2013).
Li, Y., Xiao, M. & Guo, F. The role of Sox6 and Netrin-1 in ovarian cancer cell growth, invasiveness, and angiogenesis. Tumour Biol 39, 1010428317705508, https://doi.org/10.1177/1010428317705508 (2017).
Jiang, W. et al. Identification of Sox6 as a regulator of pancreatic cancer development. J Cell Mol Med 22, 1864–1872, https://doi.org/10.1111/jcmm.13470 (2018).
Li, Y. C. et al. MicroRNA-766 targeting regulation of SOX6 expression promoted cell proliferation of human colorectal cancer. Onco Targets Ther 8, 2981–2988, https://doi.org/10.2147/OTT.S89459 (2015).
Wang, J. et al. Role of p14ARF-HDM2-p53 axis in SOX6-mediated tumor suppression. Oncogene 35, 1692–1702, https://doi.org/10.1038/onc.2015.234 (2016).
Iguchi, H. et al. SOX6 suppresses cyclin D1 promoter activity by interacting with beta-catenin and histone deacetylase 1, and its down-regulation induces pancreatic beta-cell proliferation. J Biol Chem 282, 19052–19061, https://doi.org/10.1074/jbc.M700460200 (2007).
Ueda, R., Yoshida, K., Kawakami, Y., Kawase, T. & Toda, M. Immunohistochemical analysis of SOX6 expression in human brain tumors. Brain Tumor Pathol 21, 117–120 (2004).
Dumitriu, B. et al. Sox6 is necessary for efficient erythropoiesis in adult mice under physiological and anemia-induced stress conditions. PLoS One 5, e12088, https://doi.org/10.1371/journal.pone.0012088 (2010).
Cantu, C. et al. Sox6 enhances erythroid differentiation in human erythroid progenitors. Blood 117, 3669–3679, https://doi.org/10.1182/blood-2010-04-282350 (2011).
Dumitriu, B. et al. Sox6 cell-autonomously stimulates erythroid cell survival, proliferation, and terminal maturation and is thereby an important enhancer of definitive erythropoiesis during mouse development. Blood 108, 1198–1207, https://doi.org/10.1182/blood-2006-02-004184 (2006).
Deininger, M. W., Goldman, J. M. & Melo, J. V. The molecular biology of chronic myeloid leukemia. Blood 96, 3343–3356 (2000).
Chereda, B. & Melo, J. V. Natural course and biology of CML. Ann Hematol 94(Suppl 2), S107–121, https://doi.org/10.1007/s00277-015-2325-z (2015).
Fielding, A. K. Treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia in adults: a broader range of options, improved outcomes, and more therapeutic dilemmas. Am Soc Clin Oncol Educ Book, e352-359, https://doi.org/10.14694/EdBook_AM.2015.35.e352 (2015).
Bernt, K. M. & Hunger, S. P. Current concepts in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia. Front Oncol 4, 54, https://doi.org/10.3389/fonc.2014.00054 (2014).
Levine, R. L., Pardanani, A., Tefferi, A. & Gilliland, D. G. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat Rev Cancer 7, 673–683, https://doi.org/10.1038/nrc2210 (2007).
Tang, Z. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45, W98–W102, https://doi.org/10.1093/nar/gkx247 (2017).
Iacobucci, I. et al. CDKN2A/B alterations impair prognosis in adult BCR-ABL1-positive acute lymphoblastic leukemia patients. Clin Cancer Res 17, 7413–7423, https://doi.org/10.1158/1078-0432.CCR-11-1227 (2011).
Williams, R. T., Roussel, M. F. & Sherr, C. J. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci USA 103, 6688–6693, https://doi.org/10.1073/pnas.0602030103 (2006).
Naumovski, L. et al. Philadelphia chromosome-positive acute lymphoblastic leukemia cell lines without classical breakpoint cluster region rearrangement. Cancer Res 48, 2876–2879 (1988).
Lozzio, B. B. & Lozzio, C. B. Properties of the K562 cell line derived from a patient with chronic myeloid leukemia. Int J Cancer 19, 136 (1977).
Ogura, M. et al. Establishment of a novel human megakaryoblastic leukemia cell line, MEG-01, with positive Philadelphia chromosome. Blood 66, 1384–1392 (1985).
Hong, Y., Martin, J. F., Vainchenker, W. & Erusalimsky, J. D. Inhibition of protein kinase C suppresses megakaryocytic differentiation and stimulates erythroid differentiation in HEL cells. Blood 87, 123–131 (1996).
Uozumi, K. et al. Establishment and characterization of a new human megakaryoblastic cell line (SET-2) that spontaneously matures to megakaryocytes and produces platelet-like particles. Leukemia 14, 142–152 (2000).
Fiedler, W. et al. Derivation of a new hematopoietic cell line with endothelial features from a patient with transformed myeloproliferative syndrome: a case report. Cancer 88, 344–351 (2000).
Nutt, S. L. & Kee, B. L. The transcriptional regulation of B cell lineage commitment. Immunity 26, 715–725, https://doi.org/10.1016/j.immuni.2007.05.010 (2007).
Hwang, Y. et al. Global increase in replication fork speed during ap57(KIP2)-regulated erythroid cell fate switch. Sci Adv 3, e1700298, https://doi.org/10.1126/sciadv.1700298 (2017).
Acknowledgements
This work was supported by Fondazione Cariplo grant No 2012.0517 to A.R. and by the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7/2007–2013/under REA grant agreement No 289611 (HEM_ID Project) to A.R. We thank Prof. J. Ghysdael for the gift of B-ALL cells; Dr. C. Cantù and Prof. S. Ottolenghi for critical reading of the manuscript; Dr. L. Scotti for assistance in statistical analysis.
Author information
Authors and Affiliations
Contributions
G.B. experimental work and study design; C.F. experiments discussion; S.M.L.B. manuscript discussion; A.E.R. experimental design supervision and manuscript writing. All authors participated to biological data interpretation and discussion. All authors approved the final version.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Barbarani, G., Fugazza, C., Barabino, S.M.L. et al. SOX6 blocks the proliferation of BCR-ABL1+ and JAK2V617F+ leukemic cells. Sci Rep 9, 3388 (2019). https://doi.org/10.1038/s41598-019-39926-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-39926-4
This article is cited by
-
Clinical implications and genetical insights of SOX6 expression in acute myeloid leukemia
Journal of Cancer Research and Clinical Oncology (2023)
-
RETRACTED ARTICLE: Restoring microRNA-499-5p Protects Sepsis-Induced Lung Injury Mice Via Targeting Sox6
Nanoscale Research Letters (2021)
-
Structural basis for nuclear import selectivity of pioneer transcription factor SOX2
Nature Communications (2021)
-
SOX18 promotes gastric cancer metastasis through transactivating MCAM and CCL7
Oncogene (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.