Abstract
It is unclear whether dietary lutein/zeaxanthin intake in colorectal cancer is associated with microRNA processing involved in DICER1 cleavage for messenger RNA translation. We investigated whether dietary lutein/zeaxanthin intake affects colorectal cancer risk in patients with a DICER1 rs3742330 polymorphism. In this hospital-based case-control study, we recruited 923 colorectal cancer patients and 1,846 controls based on eligibility criteria, a semiquantitative food frequency questionnaire and the DICER1 rs3742330 genotype. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated by unconditional logistic regression adjusted for confounders. The highest quartile of lutein/zeaxanthin consumption was inversely associated with a reduced colorectal cancer risk (OR, 95% CI = 0.25, 0.18–0.36). Carrying G allele (AG + GG) showed a significantly reduced colorectal cancer incidence compared with that of AA carriers (OR, 95% CI = 0.71, 0.55–0.91). Those carrying the G allele (AG + GG) along with high lutein/zeaxanthin consumption were markedly associated with a decreased colorectal cancer risk (OR, 95% CI = 0.32, 0.22–0.46, P for interaction = 0.018), particularly for rectal cancer (OR, 95% CI = 0.24, 0.15–0.39, P for interaction = 0.004), compared with that of AA carriers with low lutein/zeaxanthin intakes. In conclusion, colorectal cancer risk was related to an interactive effect between dietary lutein/zeaxanthin intake and the DICER1 rs3742330 polymorphism.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a common cancer that causes mortality worldwide, with approximately 1.4 million new cases and nearly 694,000 deaths in 20121. Cancer incidence and mortality data for 2018 indicated that CRC is one of the most common cancers in Korea and has an age-standardized incidence rate of 23.1 per 100,000 people in both sexes worldwide2. Risk factors for CRC include age (>50), history of adenomatous polyps, family history of CRC, inherited genetic risk, physical activity, obesity, smoking, alcohol consumption, and dietary habits3. A leading cause of CRC is a western-style diet, which includes low fruit and vegetable intake and is an important risk factor in developing colorectal carcinogenesis3,4.
Lutein/zeaxanthin are abundant xanthophyll carotenoids in many dark-green leafy vegetables and in egg yolks5. Lutein/zeaxanthin protect against degenerative eye conditions including age-related macular degeneration and cataracts6. Several studies have explored the association between lutein/zeaxanthin and cancer, showing that they provide potential antioxidant functions in cellular mechanisms that regulate apoptosis7,8,9, modulate gap junctional intercellular communication10, and stimulate DNA strand break repair11. Epidemiology studies have shown that lutein/zeaxanthin intake is inversely associated with breast, lung and CRC development12. However, no evidence exists to support an interaction between dietary lutein/zeaxanthin intake and genetic variants associated with a risk of developing CRC.
Many investigators have revealed the importance of microRNAs (miRNAs) in cancer initiation and progression via their effects in silencing tumor suppressive and oncogenic messenger RNAs (mRNAs)13. miRNAs are small noncoding regions that are linked to both mRNA translation and RNA silencing14. The DICER1 gene is an endonuclease with RNase III activity that cleaves pre-miRNA as part of a trans activating response RNA binding protein (TRBP) complex to generate miRNA and small interfering RNA15. DICER1 is involved in the processing necessary for miRNA biogenesis. In cytoplasmic processing, Dicer is an essential enzyme for miRNA biogenesis and maturation, enabling mRNA to repress gene expression16. The miRNAs processed by DICER1 have been reported to mediate ovarian cancer, breast cancer, and CRC through carcinogenic mechanisms such as cellular proliferation, apoptosis, and differentiation17,18. A single nucleotide polymorphism (SNP) of rs3742330, which is related to miRNA synthesis, is located in the 3′-untranslated region (UTR) of DICER1 on 14q32, and its regulation has become important in association with CRC risk19,20. However, limited evidence indicates that dietary factors may be associated with regulating DICER1 rs3742330 in CRC cases.
In this study, we analyzed DICER1 rs3742330 genotypes from DNA samples and evaluated the intake of lutein/zeaxanthin by using a validated semiquantitative food frequency questionnaire (SQFFQ) among controls and CRC patients to identify CRC etiology. This study investigated whether lutein/zeaxanthin intake shows an interaction with DICER1 rs3742330 in relation to CRC risk.
Results
General characteristics of the study participants
Table 1 shows the study subjects’ demographic characteristics. With respect to sociodemographic factors, cases were statistically associated with education level, professional occupation, income and positive first-degree family history of CRC (P < 0.001). Patients had a higher rate of ex-alcohol drinkers and physical inactivity than the controls (P < 0.001). No significant differences were observed for BMI or smoking status. The mean total energy intakes for the controls and the CRC patients were 1,689.6 ± 560.43 kcal/day and 2,2026.3 ± 533.96 kcal/day, respectively (P < 0.001). The mean lutein/zeaxanthin intake was 3.61 ± 2.91 mg/day for the controls and 2.72 ± 1.78 mg/day for the CRC patients (P < 0.001).
Association of dietary lutein/zeaxanthin intake with CRC risk
Table 2 shows the ORs and 95% CIs of CRC risk for lutein/zeaxanthin intake. Dietary lutein/zeaxanthin intake on CRC risk was compared by quartile groups. Lutein/zeaxanthin intake was strongly significantly associated with CRC risk in both crude and multivariable models adjusted for age, sex, BMI, education level, occupation, income, alcohol consumption, physical activity, first-degree family history of CRC, and total energy intake. The highest lutein/zeaxanthin intake quartile was inversely associated with CRC risk compared with that of the lowest quartile group (OR, 95% CI = 0.25, 0.18–0.36, highest versus lowest quartile, P for trend < 0.001). When stratified by anatomical subsite for CRC, the highest lutein/zeaxanthin intake showed significant inverse associations in proximal, distal, and rectal cancer compared with those of lower lutein/zeaxanthin intake (proximal colon cancer OR, 95% CI = 0.38, 0.21–0.71; distal colon cancer OR, 95% CI = 0.21, 0.12–0.35; rectal cancer OR, 95% CI = 0.26, 0.17–0.41, highest versus lowest quartile, P for trend < 0.001).
Association of DICER1 rs3742330 variant with CRC risk
Table 3 shows the ORs and 95% CIs of CRC risk according to the DICER1 rs3742330 variant. The DICER1 rs3742330 minor allele frequency was 0.44. Polymorphism in the controls was in Hardy-Weinberg equilibrium (HWE). Heterozygous AG among the cases was significantly associated with CRC risk in multivariable models adjusted for the covariates mentioned above (OR, 95% CI = 0.70, 0.53–0.91, P < 0.01). When comparing the rs3742330 allele frequencies, carrying a G allele (AG + GG) significantly decreased the risk of CRC compared with those who were AA homozygous via a dominant model (OR, 95% CI = 0.71, 0.55–0.91, P < 0.01).
Association of dietary lutein/zeaxanthin intake with CRC risk, stratified by DICER1 rs3742339 variant
Table 4 shows an interaction between lutein/zeaxanthin intake and the DICER1 rs3742330 variant with CRC risk. Lutein/zeaxanthin intake was categorized into low and high groups, and CRC was stratified by anatomic subsite. High lutein/zeaxanthin intake while carrying a G allele (AG + GG) showed a significant interaction with low CRC risk compared with that of AA homozygous patients (OR, 95% CI = 0.32, 0.22–0.46, P for interaction = 0.018, AG + GG carriers with high intake vs. AA carriers with low intake). When CRC was stratified by anatomic subsite, lutein/zeaxanthin intake was inversely associated with rectal cancer (OR, 95% CI = 0.24, 0.15–0.39, P for interaction = 0.004, AG + GG carrier with high intake vs. AA carriers with low intake).
Discussion
The present study showed an association between dietary lutein/zeaxanthin intake and CRC risk among those with the DICER1 rs3742330 genotype in a Korean population. A high intake of lutein/zeaxanthin was inversely associated with CRC risk in G allele carriers (AG + GG) compared with that in AA homozygous DICER1 rs3742330 carriers.
Previous epidemiological studies have shown conflicting data regarding the association between lutein/zeaxanthin intake and CRC risk. Case-control studies have shown that CRC and lutein/zeaxanthin intake, either individually21,22 or together23, are significantly inversely associated. In a pooled analysis of cohort studies, dietary lutein/zeaxanthin intake was associated with a slightly reduced CRC risk24, and a similar association was found for the risk of colorectal adenomas in a cohort of male health professionals25. Because dietary intake affects lutein/zeaxanthin serum levels, these levels, either alone or together, were decreased in patients with gastrointestinal cancer26, colorectal polyps27,28, and colorectal neoplasms29 compared with those in controls. In contrast, several studies reported that lutein/zeaxanthin intake was not associated with CRC risk. Six case-control studies showed that dietary intake or serum levels of either lutein only30,31,32 or lutein/zeaxanthin33,34,35 were not associated with CRC risk. Several large prospective cohort studies have supported the lack of an association between dietary lutein/zeaxanthin intake and CRC36,37. These results might be attributed to differences in the range of lutein/zeaxanthin intake, CRC status and the point that CRC examination began in the cohort study. In an in vitro study, dietary lutein inhibited colonic aberrant crypt focal development in rats, suggesting that it may help prevent colon carcinogenesis38,39. Other studies have suggested that lutein/zeaxanthin bioavailability affects carcinogenesis via its antioxidant properties by inducing apoptosis and proliferation, preventing oxygen radicals, regulating gene expression, and activating the immune response6,12,21,40. In this study, high lutein/zeaxanthin intake was strongly inversely associated with reduced CRC risk in all anatomic subsites, including the proximal and distal colon and rectum.
DICER1 plays a role in a biogenesis pathway that is known to regulate the repression of mRNA translation and trigger mRNA degradation by binding to the 3′-untranslated region (UTR) of target mRNA. Loss of function in heterozygous DICER1 germline pathogenic variants and somatic missense variants of DICER1 result in the abnormal production of miRNAs41,42,43,44,45. Abnormal miRNA expression is suspected to promote cellular carcinoma by affecting proliferation, apoptosis, mitosis, and cell-cycle progression46,47. SNPs in the 3′-UTR have been reported to contribute to the regulation of transcript stabilization48. The 3′-UTR is considered a place of pathological and polymorphic sites related to disease involving miRNA activation49. A growing body of evidence has indicated that the up- and downregulation of DICER1 are related to the development of tumorigeneses such as lung, breast, ovarian, skin, prostate cancers, and CRC via the alteration of miRNA expression17,20,50. Moreover, with respect to DICER1 related cancers, several studies have suggested that loss of function or mutation of DICER1 may affect stem cell proliferation, differentiation, and cell fate and induce embryonal or blastoma carcinomas17,51,52. In an in vivo study, inactivating Dicer1 in a mouse model showed that it functioned as a haploinsufficient tumor suppressor in retinoblastoma and promoted hepatocarcinogenesis53,54.
Several studies have explored the deregulation of DICER1, which is required for miRNA processing, as part of CRC etiology. Dicer mRNA levels were significantly increased in CRC, particularly in rectal cancer, compared with those in normal mucosa55. The downregulation of Dicer was found to be a prognostic indicator underlying tumor development in CRC56. The overexpression of Dicer was associated with shorter survival time in CRC patients57. In an in vivo study, impaired DICER1 function indicated a stemness phenotype and metastasis in colon cancer58. Taken together, evidence has shown that the interaction between DICER1 and CRC demonstrates the importance of miRNA-related SNPs (miR-SNPs), which alter miRNA levels and thereby influence mRNA transcription, in DICER1.
In recent years, functional studies have shown that miRNAs can be oncogenes or tumor suppressor genes depending on their target genes59,60. The SNP rs3742330 located in the 3′-UTR of DICER1 has been reported to be the target site of two miRNAs, miR-3622a-5p and miR-5582-5p61,62. The miR-SNP rs3742330 in DICER1 contributes to T-cell lymphoma, renal cell carcinoma, oral premalignant lesions, and CRC19,63,64,65. The variant allele of DICER1 rs3742330 is associated with CRC despite inconsistent patterns in recent literature. Zhao et al. reported that the AA allele of DICER1 rs3742330 is related to an increased CRC risk19, while Cho et al. determined that the DICER1 rs3742330 AG genotype leads to an increased risk of colon cancer but not rectal cancer66. Our findings indicated that DICER1 rs3742330 AG heterozygotes were associated with an increased risk of CRC. Moreover, when comparing the frequency of the DICER1 rs3742330 allele, carrying a G allele (AG + GG) was significantly associated with a decreased risk of CRC compared with AA homozygous carriers via a dominant model. However, when stratified by anatomical sites, DICER1 rs3742330 was significantly associated with rectal cancer. These reports and our findings suggest that DICER1 rs3742330 may impact the regulation of DICER1 expression, even if it shows a tumor-specific pattern, which requires laboratory-based functional studies.
In this study, high lutein/zeaxanthin intake while carrying a G allele (AG + GG) for DICER1 rs3742330 showed a significant interaction, thus resulting in a reduced CRC risk compared with that for low lutein/zeaxanthin intake among AA homozygous individuals. Our study suggests several potential mechanisms that account for the interaction between lutein/zeaxanthin and DICER1 rs3742330 regarding CRC risk. First, the function of DICER1, influenced by rs3742330 plays a pivotal role in CRC tumorigenesis by altering the expression of miRNA-related oncogenic pathways underlying cellular transformation, such as proliferation, apoptosis, invasion, and metastasis67. Regarding miRNA expression and CRC risk, Slattery et al. suggested that dietary and lifestyle factors related to inflammation and oxidative stress are linked to regulating miRNA expression in colorectal tissue, leading to an elevated CRC risk68. Although we did not measure miRNA expression levels by lutein/zeaxanthin intake, our study supports the notion that dietary lutein/zeaxanthin intake impacts DICER1 activity by regulating miRNA expression, thus affecting CRC risk via an underlying interaction between antioxidant effects and miRNA expression. Second, DNA damage is a possible cause of tumorigenesis69. Several studies have identified that a loss of Dicer contributes to tumorigenesis progression via DNA damage69,70. Among various nutrients related to DNA damage, Haegele et al. proposed an inverse association between plasma lutein and oxidative indices, showing that lutein affects DNA damage repair71. Lutein may influence the DNA damage repair associated with Dicer’s role in CRC risk. Third, deleting Dicer reduces T-reg cell numbers and immune pathology, indicating Dicer’s importance in immune regulation and immune cell development72,73. Several studies have reported that lutein/zeaxanthin action induces cell-mediated and humoral immune responses12,40. Because lutein/zeaxanthin exert immunological properties via the immune system, an interaction between lutein/zeaxanthin and Dicer could reduce CRC risk via an underlying immune system effect. Despite numerous studies investigating the role of the miRNA associated DICER1 in CRC risk, the interaction between DICER1 functions and other elements, such as diet and environmental factors resulting in CRC, remain undetermined.
To our knowledge, this is the first report to examine the miRNA associated DICER1 gene and dietary interaction as part of CRC etiology. Dietary lutein/zeaxanthin intake and the DICER1 rs3742330 genotype were inversely associated in CRC, providing new insight into a protective effect of lutein/zeaxanthin against CRC risk in patients carrying the G allele. However, this study has limitations that must be considered when interpreting our findings. This study was designed as a hospital-based case-control study and may have been affected by selection bias. To evaluate dietary lutein/zeaxanthin intake, we used a validated SQFFQ that may have been affected by recall bias among the participants. In addition, we analyzed the DICER1 rs3742330 genotype to determine the association between lutein/zeaxanthin intake and CRC risk. Further investigation regarding how lutein/zeaxanthin impacts miRNA associated DICER1 expression levels and miRNA processing machinery relative to CRC risk is needed to understand the associated molecular mechanisms.
The results of this study indicate that lutein/zeaxanthin affects miRNA processing by regulating DICER1, which appears to result in a lower CRC risk. Furthermore, rs3742330 may either directly or indirectly alter Dicer-mediated miRNA levels, which can influence mRNA transcription and CRC risk. We propose that future studies determine how dietary lutein/zeaxanthin intake is associated with regulating DICER1 rs3742330 and the underlying molecular mechanism in CRC risk.
Methods
Study population
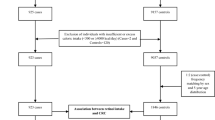
This was a hospital-based case-controlled study in a Korean population designed to detect associations between dietary intake, genetic factors, and CRC risk, which is described elsewhere4. In brief, this study by the Center for Colorectal Cancer of the National Cancer Center Korea included 1,070 patients newly diagnosed with colorectal tumors pathologically confirmed as adenocarcinoma by endoscopic biopsies who agreed to participate between August 2010 and August 2013. The 14,201 cancer-free controls, confirmed by linking to the Korea Central Cancer Registry (KCCR) database, were recruited by the Center for Cancer Prevention and Detection at the National Cancer Center Korea for a health check-up program provided by the National Health Insurance Cooperation between October 2007 and December 2017. Based on eligibility criteria, we excluded 5,189 subjects (145 cases and 5,044 controls) with an incomplete SQFFQ and 122 subjects (2 cases and 120 controls) with an implausible self-reported energy intake (<500 or >4,000 kcal/day). Of the remaining subjects, controls were matched to cases at a 1:2 ratio (cases: controls) by sex within a 5-year age range. For genetic analysis, patients with missing blood samples were excluded (Supplementary Fig. S1). This study conformed to the National Cancer Center Korea guidelines, and participants signed a written informed consent document. The Institutional Review Board of the National Cancer Center Korea approved this study (IRB Nos. NCCNCS-10-350 and NCC2015-0202).
Data collection
Demographic characteristics and lifestyle information were obtained from a structured questionnaire administered by a trained interviewer. Participants were also asked about their dietary intake using a 106-item SQFFQ that was previously determined to be reliable and valid74. Energy intake and individual nutrients were calculated using Computer Aided Nutritional Analysis Program 4.0 (CAN-PRO 4.0, The Korean Nutrition Society, Seoul, Korea). To estimate lutein/zeaxanthin intake, we used a carotenoid database developed by merging databases from the United States Department of Agriculture, the Korea functional food composition table, and literature searches75,76. From the 2,903-items in the carotenoid database, we estimated lutein/zeaxanthin intake to identify its association with CRC risk.
SNP genotyping
Genomic DNA was extracted from participants’ blood samples following the manufacturer’s instructions, using the MagAttract DNA Blood M48 Kit (Qiagen, Hilden, Germany) and BioRobot M48 automatic extraction equipment (Qiagen). SNP genotyping was performed using the MassARRAY iPLEX Gold Assay (Agena Bioscience, San Diego, CA, USA). The rs3742330 genotyping results were A > G in 1,400 controls and 700 cases.
Statistical analysis
Demographic and lifestyle characteristics were analyzed by χ2 tests for categorical variables and Student’s t-test for continuous variables. Dietary lutein/zeaxanthin intake was adjusted for total energy intake using the residual method77. To compare its association with CRC risk, dietary lutein/zeaxanthin intake was evaluated by exposure quartiles in the control intake. ORs and 95% CIs were calculated using unconditional logistic regression models, adjusting for age, sex, BMI, education level, occupation, income, alcohol consumption, physical activity, first-degree family history of CRC, and total energy intake. We analyzed CRC risk stratified by anatomical subsite (proximal colon, distal colon, and rectal cancer) using a multinomial logistic regression model. The rs3742330 was in Hardy-Weinberg equilibrium (HWE), and the genotype was categorized into dominant (AA vs. AG + GG) and recessive (AA + AG vs. GG) effect models. To examine diet-gene interaction, ORs and 95% CIs were estimated based on the rs3742330 genetic model and lutein/zeaxanthin intake categories using a multiple logistic regression model, adjusting for the aforementioned covariates. We divided the participants into two intake groups (low/high) based on median levels of control to test the interaction between lutein/zeaxanthin intake and the rs3742330 genetic model using a likelihood ratio. All analyses were estimated using SAS version 9.4 software (SAS Institute Inc., Cary, NC, USA). A significance level of less than 0.05 was used and determined with two-tailed tests.
References
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–386 (2015).
Jung, K. W., Won, Y. J., Kong, H. J. & Lee, E. S. Prediction of Cancer Incidence and Mortality in Korea, 2018. Cancer Res. Treat. 50, 317–323 (2018).
Haggar, F. A. & Boushey, R. P. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 22, 191–197 (2009).
Park, Y., Lee, J., Oh, J. H., Shin, A. & Kim, J. Dietary patterns and colorectal cancer risk in a Korean population: A case-control study. Medicine (Baltimore) 95, e3759 (2016).
Handelman, G. J., Nightingale, Z. D., Lichtenstein, A. H., Schaefer, E. J. & Blumberg, J. B. Lutein and zeaxanthin concentrations in plasma after dietary supplementation with egg yolk. Am. J. Clin. Nutr. 70, 247–251 (1999).
Ribaya-Mercado, J. D. & Blumberg, J. B. Lutein and zeaxanthin and their potential roles in disease prevention. J. Am. Coll. Nutr. 23, 567s–587s (2004).
Muller, K., Carpenter, K. L., Challis, I. R., Skepper, J. N. & Arends, M. J. Carotenoids induce apoptosis in the T-lymphoblast cell line Jurkat E6.1. Free Radic. Res. 36, 791–802 (2002).
Molnar, J. et al. Modulation of multidrug resistance and apoptosis of cancer cells by selected carotenoids. In Vivo 18, 237–244 (2004).
Chew, B. P., Brown, C. M., Park, J. S. & Mixter, P. F. Dietary lutein inhibits mouse mammary tumor growth by regulating angiogenesis and apoptosis. Anticancer Res. 23, 3333–3339 (2003).
Zhang, L. X., Cooney, R. V. & Bertram, J. S. Carotenoids enhance gap junctional communication and inhibit lipid peroxidation in C3H/10T1/2 cells: relationship to their cancer chemopreventive action. Carcinogenesis 12, 2109–2114 (1991).
Astley, S. B., Elliott, R. M., Archer, D. B. & Southon, S. Increased cellular carotenoid levels reduce the persistence of DNA single-strand breaks after oxidative challenge. Nutr. Cancer 43, 202–213 (2002).
Milani, A., Basirnejad, M., Shahbazi, S. & Bolhassani, A. Carotenoids: biochemistry, pharmacology and treatment. Br. J. Pharmacol. 174, 1290–1324 (2017).
Del Carmen Martinez-Jimenez, V., Mendez-Mancilla, A. & Patricia Portales-Perez, D. miRNAs in nutrition, obesity, and cancer: The biology of miRNAs in metabolic disorders and its relationship with cancer development. Mol. Nutr. Food Res. 62 (2018).
Huntzinger, E. & Izaurralde, E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews. Genetics 12, 99–110 (2011).
Ryan, B. M., Robles, A. I. & Harris, C. C. Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer 10, 389–402 (2010).
Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001).
Bahubeshi, A., Tischkowitz, M. & Foulkes, W. D. miRNA processing and human cancer: DICER1 cuts the mustard. Sci. Transl. Med. 3, 111ps146 (2011).
Dedes, K. J. et al. Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur. J. Cancer 47, 138–150 (2011).
Zhao, Y., Du, Y., Zhao, S. & Guo, Z. Single-nucleotide polymorphisms of microRNA processing machinery genes and risk of colorectal cancer. Onco Targets Ther. 8, 421–425 (2015).
He, J. et al. MicroRNA biogenesis pathway genes polymorphisms and cancer risk: a systematic review and meta-analysis. PeerJ 4, e2706 (2016).
Slattery, M. L. et al. Carotenoids and colon cancer. Am. J. Clin. Nutr. 71, 575–582 (2000).
Chaiter, Y. et al. Smoking attenuates the negative association between carotenoids consumption and colorectal cancer risk. Cancer Causes Control 20, 1327–1338 (2009).
Levi, F., Pasche, C., Lucchini, F. & La Vecchia, C. Selected micronutrients and colorectal cancer. a case-control study from the canton of Vaud, Switzerland. Eur. J. Cancer 36, 2115–2119 (2000).
Mannisto, S. et al. Dietary carotenoids and risk of colorectal cancer in a pooled analysis of 11 cohort studies. Am. J. Epidemiol 165, 246–255 (2007).
Jung, S. et al. Carotenoid intake and risk of colorectal adenomas in a cohort of male health professionals. Cancer Causes Control 24, 705–717 (2013).
McMillan, D. C., Sattar, N., Talwar, D., O’Reilly, D. S. & McArdle, C. S. Changes in micronutrient concentrations following anti-inflammatory treatment in patients with gastrointestinal cancer. Nutrition 16, 425–428 (2000).
Rumi, G. Jr. et al. Decrease in serum levels of vitamin A and zeaxanthin in patients with colorectal polyp. Eur. J. Gastroenterol. Hepatol. 11, 305–308 (1999).
Nair, S., Norkus, E. P., Hertan, H. & Pitchumoni, C. S. Serum and colon mucosa micronutrient antioxidants: differences between adenomatous polyp patients and controls. Am. J. Gastroenterol. 96, 3400–3405 (2001).
Okuyama, Y. et al. Inverse associations between serum concentrations of zeaxanthin and other carotenoids and colorectal neoplasm in Japanese. Int. J. Clin. Oncol. 19, 87–97 (2014).
Terry, P., Jain, M., Miller, A. B., Howe, G. R. & Rohan, T. E. Dietary carotenoid intake and colorectal cancer risk. Nutr. Cancer 42, 167–172 (2002).
Satia-Abouta, J. et al. Associations of micronutrients with colon cancer risk in African Americans and whites: results from the North Carolina Colon Cancer Study. Cancer Epidemiol. Biomarkers Prev. 12, 747–754 (2003).
Shikany, J. M. et al. Plasma carotenoids and the prevalence of adenomatous polyps of the distal colon and rectum. Am. J. Epidemiol. 145, 552–557 (1997).
Enger, S. M. et al. Dietary intake of specific carotenoids and vitamins A, C, and E, and prevalence of colorectal adenomas. Cancer Epidemiol. Biomarkers Prev. 5, 147–153 (1996).
Lu, M. S. et al. Higher intake of carotenoid is associated with a lower risk of colorectal cancer in Chinese adults: a case-control study. Eur. J. Nutr. 54, 619–628 (2015).
Huang, J. et al. Serum carotenoids and colorectal cancer risk: A case-control study in Guangdong, China. Mol. Nutr. Food Res. 61 (2017).
Malila, N. et al. Dietary and serum alpha-tocopherol, beta-carotene and retinol, and risk for colorectal cancer in male smokers. Eur. J. Clin. Nutr. 56, 615–621 (2002).
Leenders, M. et al. Plasma and dietary carotenoids and vitamins A, C and E and risk of colon and rectal cancer in the European Prospective Investigation into Cancer and Nutrition. Int. J. Cancer 135, 2930–2939 (2014).
Narisawa, T. et al. Inhibitory effects of natural carotenoids, alpha-carotene, beta-carotene, lycopene and lutein, on colonic aberrant crypt foci formation in rats. Cancer Lett. 107, 137–142 (1996).
Kim, J. M. et al. Chemopreventive effects of carotenoids and curcumins on mouse colon carcinogenesis after 1,2-dimethylhydrazine initiation. Carcinogenesis 19, 81–85 (1998).
Chew, B. P. & Park, J. S. Carotenoid action on the immune response. J. Nutr. 134, 257s–261s (2004).
Hill, D. A. et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 325, 965 (2009).
Rio Frio, T. et al. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. Jama 305, 68–77 (2011).
Schultz, K. A. et al. Ovarian sex cord-stromal tumors, pleuropulmonary blastoma and DICER1 mutations: a report from the International Pleuropulmonary Blastoma Registry. Gynecol. Oncol. 122, 246–250 (2011).
Slade, I. et al. DICER1 syndrome: clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J. Med. Genet. 48, 273–278 (2011).
Witkowski, L. et al. DICER1 hotspot mutations in non-epithelial gonadal tumours. Br. J. Cancer 109, 2744–2750 (2013).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355 (2004).
Calin, G. A. & Croce, C. M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 (2006).
Carmody, S. R. & Wente, S. R. mRNA nuclear export at a glance. J. Cell Sci. 122, 1933–1937 (2009).
Conne, B., Stutz, A. & Vassalli, J. D. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat. Med. 6, 637–641 (2000).
Shan, W. et al. Role of Dicer as a prognostic predictor for survival in cancer patients: a systematic review with a meta-analysis. Oncotarget 7, 72672–72684 (2016).
Bernstein, E. et al. Dicer is essential for mouse development. Nat. Genet. 35, 215–217 (2003).
Fukagawa, T. et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6, 784–791 (2004).
Kumar, M. S. et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 23, 2700–2704 (2009).
Sekine, S. et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology 136, 2304-2315.e2301–2304 (2009).
Stratmann, J. et al. Dicer and miRNA in relation to clinicopathological variables in colorectal cancer patients. BMC Cancer 11, 345 (2011).
Faggad, A. et al. Down-regulation of the microRNA processing enzyme Dicer is a prognostic factor in human colorectal cancer. Histopathology 61, 552–561 (2012).
Faber, C., Horst, D., Hlubek, F. & Kirchner, T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur. J. Cancer 47, 1414–1419 (2011).
Iliou, M. S. et al. Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene 33, 4003–4015 (2014).
Peng, Y. & Croce, C. M. The role of MicroRNAs in human cancer. Signal transduction and targeted therapy 1, 15004 (2016).
Adams, B. D., Kasinski, A. L. & Slack, F. J. Aberrant regulation and function of microRNAs in cancer. Curr. Biol. 24, R762–776 (2014).
Persson, H. et al. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 71, 78–86 (2011).
Friedlander, M. R., Mackowiak, S. D., Li, N., Chen, W. & Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52 (2012).
Horikawa, Y. et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res. 14, 7956–7962 (2008).
Li, X., Tian, X., Zhang, B., Zhang, Y. & Chen, J. Variation in dicer gene is associated with increased survival in T-cell lymphoma. PLoS One 7, e51640 (2012).
Clague, J. et al. Genetic variation in MicroRNA genes and risk of oral premalignant lesions. Mol. Carcinog. 49, 183–189 (2010).
Cho, S. H. et al. 3′-UTR Polymorphisms in the MiRNA Machinery Genes DROSHA, DICER1, RAN, and XPO5 Are Associated with Colorectal Cancer Risk in a Korean Population. PLoS One 10, e0131125 (2015).
Strubberg, A. M. & Madison, B. B. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis. Model. Mech. 10, 197–214 (2017).
Slattery, M. L., Herrick, J. S., Mullany, L. E., Stevens, J. R. & Wolff, R. K. Diet and lifestyle factors associated with miRNA expression in colorectal tissue. Pharmgenomics Pers. Med. 10, 1–16 (2017).
Tang, K. F. & Ren, H. The role of dicer in DNA damage repair. Int. J. Mol. Sci. 13, 16769–16778 (2012).
Kumar, M. S., Lu, J., Mercer, K. L., Golub, T. R. & Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39, 673–677 (2007).
Haegele, A. D. et al. Plasma xanthophyll carotenoids correlate inversely with indices of oxidative DNA damage and lipid peroxidation. Cancer Epidemiol. Biomarkers Prev. 9, 421–425 (2000).
Cobb, B. S. et al. A role for Dicer in immune regulation. J. Exp. Med. 203, 2519–2527 (2006).
Devasthanam, A. S. & Tomasi, T. B. Dicer in immune cell development and function. Immunol. Invest. 43, 182–195 (2014).
Ahn, Y. et al. Validation and reproducibility of food frequency questionnaire for Korean genome epidemiologic study. Eur. J. Clin. Nutr. 61, 1435–1441 (2007).
Regu, G. M. et al. Association between Dietary Carotenoid Intake and Bone Mineral Density in Korean Adults Aged 30–75 Years Using Data from the Fourth and Fifth Korean National Health and Nutrition Examination Surveys (2008–2011). Nutrients 9 (2017).
Kim, J. H. Dietary Carotenoids Intake and Risk of Gastric Cancer: a Case-control Study in Korea Master thesis, National Cancer Center Graduate School of Cancer Science and Policy (2018).
Willett, W. C., Howe, G. R. & Kushi, L. H. Adjustment for total energy intake in epidemiologic studies. Am. J. Clin. Nutr. 65, 1220S–1228S; discussion 1229S–1231S (1997).
Acknowledgements
We gratefully acknowledge the efforts of those who have contributed in this study. This study was supported by grants from National Cancer Center Korea (NO: 1710882, 1810090).
Author information
Authors and Affiliations
Contributions
The authors’ responsibilities were as follows: J.M.K. and J.S.K. conducted the research design, statistical analysis, and writing of this paper. J.L., J.H.O., H.J.C., D.K.S., O.K., and A.S. recruited the study participants, collected the data, and conducted the research. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, J., Lee, J., Oh, J.H. et al. Dietary Lutein Plus Zeaxanthin Intake and DICER1 rs3742330 A > G Polymorphism Relative to Colorectal Cancer Risk. Sci Rep 9, 3406 (2019). https://doi.org/10.1038/s41598-019-39747-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-39747-5
This article is cited by
-
Lutein, violaxanthin, and zeaxanthin spectrophotometric quantification: A machine learning approach
Journal of Applied Phycology (2023)
-
Niacin, lutein and zeaxanthin and physical activity have an impact on Charlson comorbidity index using zero-inflated negative binomial regression model: National Health and Nutrition Examination Survey 2013–2014
BMC Public Health (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.