Abstract
Identification of cancer prognostic genes is important in that it can lead to accurate outcome prediction and better therapeutic trials for cancer patients. Many computational approaches have been proposed to achieve this goal; however, there is room for improvement. Recent developments in deep learning techniques can aid in the identification of better prognostic genes and more accurate outcome prediction, but one of the main problems in the adoption of deep learning for this purpose is that data from cancer patients have too many dimensions, while the number of samples is relatively small. In this study, we propose a novel network-based deep learning method to identify prognostic gene signatures via distributed gene representations generated by G2Vec, which is a modified Word2Vec model originally used for natural language processing. We applied the proposed method to five cancer types including liver cancer and showed that G2Vec outperformed extant feature selection methods, especially for small number of samples. Moreover, biomarkers identified by G2Vec was useful to find significant prognostic gene modules associated with hepatocellular carcinoma.
Similar content being viewed by others
Introduction
Accurate identification of cancer prognostic genes is important in that it can lead to improvement in the accuracy of outcome prediction and better therapeutic trials for cancer patients. Many computational approaches have applied various statistical or machine learning methods to gene expression profiles to achieve this goal. For instance, Emura et al. proposed a copula-based application of Cox regression for selecting prognostic markers from gene expression data1, and Sun et al. exploited support vector machine, which is a renowned machine learning model, to predict cancer prognosis2. These approaches successfully identify individual genes that can be used for cancer prognosis, but more recent studies proposed to exploit biological network data to identify group of interacting genes that can increase the accuracy in the prediction of cancer prognosis. For example, Langfelder and Horvath explored a gene module in a weighted correlation network and selected prognostic genes with a trait-based gene significance measure3. Wu and Stein derived prognostic genes using the Markov Cluster Algorithm and supervised principal component analysis4. Google’s PageRank algorithm, which ranks websites by counting the number and quality of links to a page, was utilized to evaluate the importance of a gene in prognosis using the gene network5. Development of these new methods continues to improve the accuracy of outcome prediction in several cancer types. Moreover, the resulting gene module or sub-networks of genes enables better understanding of molecular mechanisms of tumor progression. However, we paid attention to the possibility that deep learning models permit more accurate prediction of cancer outcome and identification of gene networks with richer information
The recent development of deep learning has been remarkable, and it has outperformed traditional machine learning methods, especially in image and natural language processing6,7,8. A variety of deep learning models have already been employed in many bioinformatics domains, such as protein structure prediction, biomedical imaging, and biomedical signal processing6. Esteva et al. trained a Convolutional Neural Network (CNN) model using about 130,000 skin images and was able to diagnose skin cancer as accurately as a dermatologist7. A Recurrent Neural Networks (RNN) is renowned for analyzing enormous medical documents, and it was utilized to precisely extract medical information from unstructured text of electronic health record notes8. In view of the previous results, the application of the deep learning models is also expected to demonstrate excellent performance in analyzing omics data including gene expression data for identification of better prognostic genes and more accurate outcome prediction.
One of the main problems regarding the adoption of deep learning for this purpose is that, different from image or document data, cancer patient data have too many dimensions for a relatively small number of samples. Exploiting additional network data can be an efficient solution to this problem, because they can explain dependencies between genes, which can result in a similar effect to reduction of a dimension. However, it is impossible to use biological networks to learn cancer patient data using the existing DNN (Deep Neural Networks), CNN and RNN based methods without great modification.
To address this problem, we took inspiration from Word2Vec9 and Node2Vec10. Word2Vec was developed to measure syntactic and semantic word similarities among words9. There are two models, the continuous bag-of-words (CBOW) and the skip-gram11. Both provide a word vector representation that is trained from a large dataset for vocabulary and context, so the similarity between words can be easily computed by using vector algebraic operations. Node2Vec, which is a variation of Word2Vec, regards a random path generated by a random walk as a sentence or set of words. Hence, Node2Vec can capture feature representations of nodes in networks in the similar way as Word2Vec models generate vector representation of words10. Node2Vec has been utilized to predict protein-protein interactions10 or to identify disease-associated single nucleotide polymorphisms (SNPs) by using feature representations learned from networks12.
In this study, we propose a novel network-based deep learning method, called G2Vec, to identify prognostic gene signatures. G2Vec is a modified CBOW model and provides distributed gene representations by learning gene correlation networks regarding good and poor prognosis groups made from the functional interaction (FI) network13. These random paths generated from correlation networks are thought of as sentences in the context of the cancer prognosis. G2Vec is trained by predicting whether an input path was generated from a good or poor network and provides learned distributed gene representations for identifying prognostic markers. Then we computed gene scores using the gene vector representations and identified 100 biomarker genes with high scores.
We applied the proposed gene selection procedure to gene expression data for five cancer types: bladder urothelial carcinoma (BLCA), breast invasive cancer (BRCA), cervical and endocervical cancers (CESC), acute myeloid leukemia (LAML), and liver hepatocellular carcinoma (LIHC), from The Cancer Genome Atlas (TCGA). For all cancer types, we observed two clearly distinguished gene groups that represent good and poor prognosis groups when visualizing distributed gene representations, and biomarkers selected from those gene groups showed higher prediction accuracy than genes identified by existing feature selection methods when the random forest classifier was applied14. Gene-annotation enrichment analysis on the LIHC dataset resulted in gene modules for biological functions associated with liver cancer prognosis. These gene modules include novel prognostic biomarkers, INPP4B, RUVBL1, and HDAC1, with their roles in the progression of liver cancer. Consequently, we suggest G2Vec as an effective tool for studying cancer data, and this may form a foundation for application of deep learning for exploiting gene network data.
Results
Description of data sets
We downloaded five high-throughput sequencing datasets and the corresponding clinical information for BLAC, BRCA, CESC, LAML, and LIHC from the Broad Institute GDAC Firehose15. We used the RSEM genes normalized results (level 3) and normalized the expression values using log2 transformation after adding an offset of 1. For each cancer type, we set a criterion of survival times to manage a classification problem. The outcome of patients who survived longer than the criterion and had no death events was defined as good prognosis. In contrast, we defined the prognosis for patients with reported deaths within the criterion as poor prognosis. Table 1 shows a summary of gene expression datasets.
We also downloaded FI network data from the Reactome database16 accessed in April 2017. The FI network is derived from curated pathways, protein-protein interactions, gene co-expression, GO annotations, and text-mined protein interactions13. After filtering the predicted edges from the network, the number of interactions in the FI network is 298,799. We also downloaded two other gene networks, BioGrid17 and HumanNet18. The number of edges in BioGrid and HumanNet are 416,692 and 474,620, respectively.
Visualization of distributed gene representations
G2Vec provides numeric representations of genes, which enable quantitative analysis for genes associated with cancer prognosis. We first visualized distributed gene representation to design an effective approach for identification of prognostic gene markers by t-distributed stochastic neighbor embedding (t-SNE)19, which is a broadly used dimensional reduction and visualization method. Figure 1 shows all distributed gene representations computed by G2Vec. In all cancer types, we confirmed that there were three distinct gene groups, a large central vector group and two gene vector groups separated from the center. The gene vectors in the center were initial random vectors that are not updated through the training process. Gene vectors retain initial vectors because the corresponding genes were not contained in any random paths. Two trained gene groups appeared to be related to the good and poor prognosis groups, respectively. Each gene group was composed mainly of genes whose frequency in the paths generated from good or poor outcome group (Fig. 1). Genes having same label tended to be agglomerated except for yellow labeled genes, so we thought that those genes may have important information for distinguishing two prognosis groups. Moreover, these two groups tended to be far from the center of the initial vectors. Gene vectors of the two groups were tuned to discriminate between two correlation networks comprising good and poor prognosis groups through the optimization of the deep learning model in G2Vec. Hence, we assumed that the farther a gene vector is from the center, the more relevant the corresponding gene is to cancer prognosis. We applied K-means clustering algorithm with K = 3 to distributed gene representations and found the initial random vector group and two interesting gene groups that are distinguishable from the initial random vector group (Supplementary Fig. S1). We named the two distinguishable vector groups good L-group and poor L-group, respectively.
Scatter plots of distributed gene representations with t-SNE for 5 cancer types. (A) BLCA, (B) BRCA, (C) CESC, (D) LAML, and (E) LIHC; blue and red points represent genes more frequently occurred in the paths of good and poor prognosis groups, respectively. Yellow points indicate genes whose numbers of occurrence in two prognosis groups were equal. Green points represent genes with no occurrence in any path.
Identification of biomarkers and outcome prediction
Using distributed gene representations, we scored genes to identify markers for predicting patient outcomes. We selected prognostic genes from each L-group using the d-score, which is the distance between vectors and the center, and the t-score, which measures the difference in gene expression levels between good and poor prognosis groups. We evaluated the power of biomarkers by comparing the prediction accuracy with three existing feature selection methods, NCPR5, WuStein4, and WGCNA3. In addition, we also selected biomarkers with high occurrence frequency in the generated paths. We named this approach Diff-Freq and selected top 50 genes from each good and poor prognosis group with the difference of occurrence frequencies in the paths, using Diff-Freq. We applied these feature selection methods to five sets of gene expression data from TCGA and predicted patient outcomes using the random forest classifier. We conducted 10-fold cross validation and computed the prediction accuracy in terms of the area under the receiver operating curve (AUC-ROC). The results of the prediction accuracy are shown in Fig. 2. We confirmed that the performance of G2Vec was higher than the others (0.009–0.049) in most cancer types, with the exception of BRCA. In BRCA, G2Vec was not the best, but it still showed good accuracy.
We also applied G2Vec to BioGrid and HumanNet in addition to the FI network. Supplementary Fig. S2 showed the AUC-ROC values using FI were slightly higher than other networks, but differences were not significant in all cancer types, which means that G2Vec works for any gene network without bias. Following experiments were conducted with FI network.
Effective sample size for G2Vec
Next, we checked the effective sample size of G2Vec to show that G2Vec is effective for small number of gene expression profiles. For each cancer type, we generated 10 sets of training and test data. Training data is composed of 10, 20, or 30 samples that are randomly chosen from each prognosis group, and the remainder samples were used as test data. In the similar way as in the previous experiments, we computed prediction accuracy of G2Vec and the extant methods on generated small datasets. Figure 3 shows that G2Vec outperformed the existing methods in most cases. When using 60 samples, G2Vec showed similar accuracy to the cases when using whole samples. Taken together, we can say that G2Vec has strength in handling a small number of samples.
Identification of prognostic modules from gene networks
Distributed gene representations can be used to find significant prognostic gene modules associated with cancer prognosis. We identified prognostic modules associated with liver cancer using 100 biomarkers identified by G2Vec (Supplementary Table S1) and 9 known oncogenes associated with hepatocarcinoma that were downloaded from the IntOGen database20. We first created two subnetworks from the entire FI network to investigate good and poor L-groups. All edges having at least one of the biomarkers or oncogenes were collected, and genes linked with only one among the biomarkers and oncogenes were removed. The good L-group and poor L-group subnetworks consisted of 280 and 198 genes, respectively (Supplementary Table S2); these subnetworks are shown in Fig. 4. Next, we carried out gene-annotation enrichment analysis using DAVID 6.8 database21 to explore biological functions or pathways in each subnetwork and identified 104 and 60 significant Gene Ontology (GO) terms and KEGG pathways (Supplementary Table S3) from the good L-group and poor L-groups, respectively. We then conducted the log-rank test22 on biomarkers contained in these GO terms or pathways to identify prognostic gene modules. For the other cancer types, the lists of GO terms and KEGG pathways were provided in Supplementary Tables S4–S7.
Subnetworks with biomarkers and oncogenes for LIHC. (A) Good L-group specific subnetwork of FI network; (B) Poor L-group specific subnetwork of FI network; Red, green, and blue genes stand for oncogenes, biomarkers, and the others, respectively. Triangle and V genes represent genes that are expressed higher and lower in poor prognosis group than in good prognosis group, respectively.
Using the aforementioned procedure, we found two significant prognostic modules, GO:0043647~inositol phosphate metabolic process and GO:1904837~beta-catenin-TCF complex assembly from the good L-group and poor L-group subnetworks, respectively. These modules and their results in terms of survival analysis are shown in Fig. 5. The Kaplan-Meier plots in Fig. 5 show that the high expressed inositol phosphate module and the low expressed beta-catenin-TCF complex represent good prognosis in patients with liver cancer. Inositol phosphates are known to play crucial roles in various cellular functions, such as cell growth, apoptosis, and cell differentiation. In particular, the INPP4B gene, identified as a biomarker of LIHC by G2Vec, has been studied as a tumor suppressor in various cancer types, such as breast, ovary, and prostate cancers23. The overexpression of INPP4B suppresses the PI3K/AKT signaling pathway and results in reduced tumor growth, which appears to be associated with PTEN24. This result may support our finding that low expression of inositol phosphates with INPP4B is associated with poor prognosis of LIHC. In addition, the beta-catenin-TCF complex has been broadly researched as a therapeutic target for colorectal and gastrointestinal cancer25,26. In the liver, the beta-catenin-TCF complex is known to suppress HNF4-alpha, which plays an essential role in development and organogenesis27, and HNF4-alpha is suggested to be an inhibitor of hepatocellular carcinoma28. These results may suggest that overexpression of the beta-catenin-TCF complex is associated with poor prognosis of liver cancer. Thus, we expect that the inositol phosphate and beta-catenin-TCF complexes would play key roles in cancer prognosis.
Significant prognostic gene modules. (A) Subnetwork of GO:0043647 from good L-group specific subnetwork; (B) Kaplan-Meier graph of GO:0043647; (C) Subnetwork of GO:1904837 from poor L-group specific subnetwork; (D) Kaplan-Meier graph of GO:1904837; In the Kaplan-Meier graph, High expression indicates the patient group whose INPP4B (or the mean of RUVBL1 and HDAC1) expression value is higher than the average of expression of the gene on the whole patients. Low expression represents the patient group whose gene expression value is lower than the average. P-values denoted in Kaplan-Meier plots are computed with log-rank test.
Discussion
In this paper, we propose a network-based deep learning model called G2Vec to identify prognostic biomarkers and gene modules. G2Vec showed significantly higher prediction accuracy for patient outcomes than existing gene selection methods. In addition, we found significant prognostic modules associated with hepatocellular carcinoma using biomarkers identified by G2Vec.
While general deep learning models require a large number of training samples, we showed that G2Vec is effective for small number of training samples (Fig. 3). This is because G2Vec generates many random paths from a correlation network. In our experiments, G2Vec generated about 43,000 paths (Supplementary Fig. S3), which is the number of inputs enough large to train a deep learning model, from gene expression data with sample size of about 130.
G2Vec has a couple of issues in training process. First, optimal parameters of G2Vec were obtained by heuristic approach. G2Vec has three critical parameters: length of random paths, size of embedded representations, and learning rate. Optimal parameters are ones that results in highest prediction accuracy of modified CBOW. The optimal maximum length of a random path, the size of projection layer, and learning rate were set to 80, 128, and 0.005, respectively (Supplementary Fig. S4). Although these values showed the best accuracy in our experiment, optimal parameters can be changed on different datasets. Second, we exploited early stopping to avoid overfitting29. In all cancer types, the modified CBOW was trained within 30 epochs, which may be too small to learn network structure completely. However, we confirmed that G2Vec showed good performance using the small epochs in Figs 2 and 3, which means that the number of epoch is sufficient. In addition, the small number guarantees fast processing time, which is another merit of G2Vec.
G2Vec is a deep learning-based method that learns the network structure, and we used it to analyze gene expression data. However, G2Vec can be extended for another omics data such as SNPs, copy number variation, or DNA methylation data, because G2Vec can be applied to any kinds of data that can be used to make correlation networks. In the future, we plan to apply G2Vec to multiple omics datasets and develop integrative analysis method for cancer research.
Methods
Overview of gene selection and validation process
The proposed gene selection method consists of three steps, generating distributed gene representations, finding L-groups, and computing gene scores. Then prognosis is predicted through 10-fold cross validation using the random forest classifier. Figure 6 shows an overview of biomarker selection and prognosis prediction. G2vec was implemented in Python3 with tensorflow and scikit-learn modules. The source code is freely available on GitHub at https://github.com/mathcom/G2Vec.
Distributed gene representations
G2Vec is a modified CBOW model to compute distributed representations of genes. From each gene correlation network from the good and poor prognosis groups, random paths are generated using a random walk algorithm. These random paths are thought of as sentences from the context of good or poor outcome groups and used for learning distributed gene representations that distinguish good and poor groups by G2Vec.
G2Vec has three steps, constructing correlation networks, generating random paths, and training a neural network. An overview of G2Vec is shown in Fig. 7.
First, we created two correlation networks associated with good and poor outcomes from the FI network and gene expression data. For each outcome group, we computed an absolute value of Pearson’s correlation coefficient (PCC) between expression levels of two genes linked on the FI network, and the values were used as a weight of the interaction. Some interactions with weight <0.5 were removed to make the difference between the two correlation networks clearer.
Next, we applied the random walk algorithm to each weighted correlation network, with three constraints. First, we never revisited a node that was passed. Second, a next destination was determined proportionally to the weight of edges. Lastly, we stopped when reaching a dead end or when the length of a path is equal to a pre-defined maximum length of the path. In this study, the maximum length of a path was set to 80. We departed from all genes 10 times, and paths were collected to learn a neural network. We gathered numerous random paths from correlation networks of good and poor prognosis groups and removed a redundant path generated in both networks. Among the filtered paths, 80% and 20% are used as training and validation datasets, respectively.
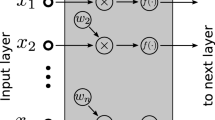
Last, we trained a modified CBOW neural network model with one hidden layer, called a projection layer. A path was represented as a binary vector whose size was equal to the number of genes. The value of a binary vector indicates whether a gene is contained in a random path or not. A neural network predicts whether an input path is generated from good or poor prognosis groups. The neural network is trained using the cross entropy objective function and Adam optimization algorithm30. Early stopping with validation set was used to avoid overfitting. We heuristically set the optimal parameters of a neural network, such as number of projection neurons and learning rate (Supplementary Fig. S4). In this study, we used 128 projection neurons and learning rate of 0.005. The trained weights between the input and projection layer (WN×M in Fig. 7) were used as gene vector representations.
L-group and gene scores
Distributed gene representations generated by G2Vec were used to group genes and to compute gene scores for identification of prognostic biomarkers. Gene vectors formed two gene groups associated with either good or poor outcome groups (Fig. 1). These groups named L-groups could be detected with K-means clustering algorithm (Supplementary Fig. S1). We then selected prognostic biomarkers from each L-group with gene scores. A gene score was defined by the means of d-scores and t-scores. A d-score is the Euclidean distance between a gene vector and the center of initial gene vectors (zero vector). A t-score is the absolute value of t-statistics measuring the difference of gene expression levels between good and poor outcomes. Both scores were normalized from 0 to 1 by min-max transformation. We selected 50 genes with high gene scores from each L-group, resulting in 100 biomarkers. For each fold, 100 biomarkers were identified using training data and validated with test data using the random forest classifier.
References
Emura, T. & Chen, Y. H. Gene selection for survival data under dependent censoring: A copula-based approach. Statistical methods in medical research 25, 2840–2857, https://doi.org/10.1177/0962280214533378 (2016).
Sun, B. Y., Zhu, Z. H., Li, J. & Linghu, B. Combined feature selection and cancer prognosis using support vector machine regression. IEEE/ACM transactions on computational biology and bioinformatics 8, 1671–1677, https://doi.org/10.1109/tcbb.2010.119 (2011).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559, https://doi.org/10.1186/1471-2105-9-559 (2008).
Wu, G. & Stein, L. A network module-based method for identifying cancer prognostic signatures. Genome biology 13, R112 (2012).
Choi, J., Park, S., Yoon, Y. & Ahn, J. Improved prediction of breast cancer outcome by identifying heterogeneous biomarkers. Bioinformatics (Oxford, England) 33, 3619–3626, https://doi.org/10.1093/bioinformatics/btx487 (2017).
Min, S., Lee, B. & Yoon, S. Deep learning in bioinformatics. Briefings in bioinformatics 18, 851–869, https://doi.org/10.1093/bib/bbw068 (2017).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118, https://doi.org/10.1038/nature21056 (2017).
Jagannatha, A. N. & Yu, H. Bidirectional RNN for Medical Event Detection in Electronic Health Records. Proceedings of the conference. Association for Computational Linguistics. North American Chapter. Meeting 2016, 473–482 (2016).
Mikolov, T., Sutskever, I., Chen, K., Corrado, G. S. & Dean, J. Distributed representations of words and phrases and their compositionality. In Advances in neural information processing systems, 3111–3119 (2013).
Grover, A. & Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD international conference on Knowledge discovery and data mining, 855–864 (2016).
Mikolov, T., Chen, K., Corrado, G. & Dean, J. Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781 (2013).
Wu, M. et al. Integrating embeddings of multiple gene networks to prioritize complex disease-associated genes. In 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 208–215 (2017).
Wu, G., Feng, X. & Stein, L. A human functional protein interaction network and its application to cancer data analysis. Genome biology 11, R53, https://doi.org/10.1186/gb-2010-11-5-r53 (2010).
Breiman, L. Random forests. Machine learning 45, 5–32 (2001).
Broad Institute TCGA Genome Data Analysis Center. Analysis-ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. Broad Institute of MIT and Harvard. Dataset, https://doi.org/10.7908/C11G0KM9 (2016).
Fabregat, A. et al. The Reactome pathway Knowledgebase. Nucleic acids research 44, D481–487, https://doi.org/10.1093/nar/gkv1351 (2016).
Chatr-Aryamontri, A. et al. The BioGRID interaction database: 2017 update. Nucleic acids research 45, D369–D379 (2017).
Lee, I., Blom, U. M., Wang, P. I., Shim, J. E. & Marcotte, E. M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome research, gr. 118992.118110 (2011).
Maaten, L. V. D. & Hinton, G. Visualizing data using t-SNE. Journal of machine learning research 9, 2579–2605 (2008).
Rubio-Perez, C. et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer cell 27, 382–396 (2015).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research 37, 1–13, https://doi.org/10.1093/nar/gkn923 (2009).
Bland, J. M. & Altman, D. G. The logrank test. BMJ (Clinical research ed.) 328, 1073, https://doi.org/10.1136/bmj.328.7447.1073 (2004).
Kofuji, S. et al. INPP4B is a PtdIns (3, 4, 5) P3 phosphatase that can act as a tumor suppressor. Cancer discovery 5, 730–739 (2015).
Gewinner, C. et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer cell 16, 115–125 (2009).
Chen, H.-J., Hsu, L.-S., Shia, Y.-T., Lin, M.-W. & Lin, C.-M. The β-catenin/TCF complex as a novel target of resveratrol in the Wnt/β-catenin signaling pathway. Biochemical pharmacology 84, 1143–1153 (2012).
Kolligs, F. T., Bommer, G. & Göke, B. Wnt/beta-catenin/tcf signaling: a critical pathway in gastrointestinal tumorigenesis. Digestion 66, 131–144 (2002).
Gougelet, A. et al. T‐cell factor 4 and β‐catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology (Baltimore, Md.) 59, 2344–2357 (2014).
Ning, B.-F. et al. Hepatocyte nuclear factor 4α suppresses the development of hepatocellular carcinoma. Cancer research 70, 7640–7651 (2010).
Caruana, R., Lawrence, S. & Giles, C. L. Overfitting in neural nets: Backpropagation. conjugate gradient, and early stopping. In Advances in neural information processing systems. 402–408 (2001).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014).
Acknowledgements
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education [NRF-2016R1D1A1B03934135] and Incheon National University (International Cooperative) Research Grant in 2016 [2016-1913].
Author information
Authors and Affiliations
Contributions
J.C. and J.A. designed the research. J.C., I.O. and S.S. carried out the experiments, wrote the program, and analyzed the data. J.C. and J.A. wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Choi, J., Oh, I., Seo, S. et al. G2Vec: Distributed gene representations for identification of cancer prognostic genes. Sci Rep 8, 13729 (2018). https://doi.org/10.1038/s41598-018-32180-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-32180-0
Keywords
This article is cited by
-
Learning functional properties of proteins with language models
Nature Machine Intelligence (2022)
-
Juxtapose: a gene-embedding approach for comparing co-expression networks
BMC Bioinformatics (2021)
-
GVES: machine learning model for identification of prognostic genes with a small dataset
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.