Abstract
Understanding structural controllability of a complex network requires to identify a Minimum Input nodes Set (MIS) of the network. Finding an MIS is known to be equivalent to computing a maximum matching of the network, where the unmatched nodes constitute an MIS. However, maximum matching is often not unique for a network, and finding all possible input nodes, the union of all MISs, may provide deep insights to the controllability of the network. Here we present an efficient enumerative algorithm for the problem. The main idea is to modify a maximum matching algorithm to make it efficient for finding all possible input nodes by computing only one MIS. The algorithm can also output a set of substituting nodes for each input node in the MIS, so that any node in the set can replace the latter. We rigorously proved the correctness of the new algorithm and evaluated its performance on synthetic and large real networks. The experimental results showed that the new algorithm ran several orders of magnitude faster than an existing method on large real networks.
Similar content being viewed by others
Introduction
Controlling complex networks1,2,3 is of great importance in many applications, such as social, biological and technical networks. For example, it has been shown that understanding network controllability can help identify genes responding to viral infection4 and genes related to cancer5, as well as assist analyzing metabolic process6.
A network is said to be controllable if it can be driven from any initial state to a desirable state in finite steps by exerting external control signals on some selected nodes1, which are called driver nodes7, input nodes8 or control nodes9. The controllability of a network can be determined by Kalman’s controllability rank condition1 if the weight of each edge is known, or Lin’s structural controllability theory10 if only a zero or non-zero value of each edge’s weight is known. As edge weights of many real networks cannot be precisely measured and are often unknown, the structural controllability theory has been widely adopted recently to analyze biological systems, such as protein interaction network4, 5, 11, signaling pathway network12, and gene regulatory network13, 14.
Based on the structural controllability theory, one approach7,8,9, 15, 16 assumes that a node can control only one of its outgoing neighbor. Therefore, input nodes of a network can be inferred by finding a maximum matching of the network, which is consisted of the set of maximum edges that do not share nodes3. The unmatched nodes related to a maximum matching constitute a Minimum Input nodes Set, or MIS. However, this approach can be only applied to directed networks. Another approach5, 17, 18 assumes that a node can control all of its outgoing edges independently, that is, a node can output different signals for each edge. Under this assumption, a minimum node dominate set (MDS) can be used to control the network18. This approach may be more reasonable for artificial networks and can be applied to undirected networks.
Based on above frameworks, extensive works have been done about control principles of complex networks. It has been shown that the size of an MIS is closely related to the node degree distribution of the network7. Interestingly, the fraction of input nodes is primarily determined by the nodes of low in- and out-degrees16. The concepts of input nodes and MIS are also extensively used in analyzing many biological networks, e.g., identifying important proteins in biological networks4, 5, analyzing interbank networks19, and increasing the effectiveness of selective modulation of brain networks20.
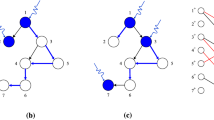
Unfortunately, maximum matching is not unique for most networks21 (Fig. 1), so there may exist numerous MISs. Although these MISs have the same size, they may have different input nodes. We call a node in an MIS a possible input node. Apparently, all possible input nodes are the union of all MISs. It is important to find all possible input nodes for understanding the controllability of a complex network. For example, finding all possible input nodes may help understand the roles of nodes in control systems22, design suitable MISs under different constraints8, and identify critical genes on signaling pathways14. Finding all possible input nodes is essentially an enumeration problem. While enumeration problems have been extensively studied, such as for enumerating all maximum matchings23, 24 or all maximally-matchable edges25, there are very few works on how to find all unmatched nodes. To solve this problem, a previous approach22 first computes a maximum matching and then assess if an unmatched node is a possible input node by removing it to test if its removal may result in a larger maximum matching. The computational complexity of finding a maximum matching is O(N 1/2 L) and the evaluation process is O(NL) on a network of N nodes and L edges, for a total complexity of O(NL) for this method.
We developed an efficient algorithm for finding all possible input nodes of a network. We proved that all possible input nodes could be identified by a simple modification to a maximum matching algorithm. The complexity of our algorithm is O(N 1/2 L), which is the same as the complexity of the maximum matching algorithm. Because our algorithm does not need to evaluate every node of the network, it runs several orders of magnitude faster than the previous method22 on large networks. Furthermore, our algorithm can also output the substituting nodes set for each input nodes. Because some input nodes may not be suitable to be inputted control signals due to some economic or technical constraints, these substituting nodes can be used to replace the original input nodes and obtain new MISs with some input nodes replaced.
Method
Consider a directed network G(V, E) over a set of nodes V and a set of edges E. To find an MIS of a directed network G(V, E), we first convert G(V, E) to an equivalent undirected bipartite graph B(V in , V out , E) (Fig. 2A,B). The bipartite graph is built by splitting the node set V into two node sets V in and V out, where a node n in G is converted to two nodes n in and n out in B, and nodes n in and n out are, respectively, connected to the in-edges and out-edges of node n.
Now consider maximum matching. A matching is a set of edges that share no common node. A node is called a matched node if it is connected to a matching edge, or unmatched node, otherwise. A matching with the maximum number of edges is called a maximum matching. In an undirected bipartite graph, an alternating path is a path whose edges are alternate in and out matching. An augmenting path is an alternating path whose two end nodes are unmatched nodes. Based on the Berge theorem26, a matching M * is a maximum matching if there is no augmenting path in B(v 1 , v 2 , E) with respect to M *. The input nodes are the unmatched nodes in V in corresponding to a maximum matching of bipartite graph B(V in, V out, E). The unmatched nodes in V in corresponding to any maximum matching form an MIS of G. If there exists a perfect matching in the network, the MIS can be any node of the network based on the Minimum Input Theorem7.
Because maximum matching is not unique for most networks, there may exist many MISs. The union of all MISs contains all possible input nodes. We now show that to find all possible input nodes of a network, we only need to compute one maximum matching without enumerating all MISs or evaluating all matched nodes as done before in ref. 22.
Theorem 1: Given a directed network G and a maximum matching M of G, a node n is a possible input node if it satisfies one of the following two conditions:
-
1.
Node n is an input node related to M;
-
2.
The in-node n in of the bipartite graph B can be reached from an input node m in related to M through an alternating path p nm .
Proof: It is sufficient to prove condition 2.
Sufficiency. Suppose that node n satisfies condition 2. Apparently, the length of p nm must be even because both node n in and m in are in the set V in of bipartite graph B. Therefore, the alternating path p nm must start with an unmatched edge connected to m in and end with a matched edge connected to node n in. Change the types of all edges of p nm , i.e., change the matched edges to unmatched and the unmatched edges to matched, then the new path p’ nm is still an alternating path. Consequently, we get a new maximum matching M’. Clearly, the node n in is not matched by M’ and m in is matched by M’. Therefore, node n is an input node of MIS D’ = D-{m} + {n}.
Necessity. Let node n in be matched in M and be unreachable by any input node related to M. Suppose that node n in is not matched by a maximum matching M’. Node n in must have at least one in-edge because it is matched by M. Therefore, there must be an alternating path p nm related to M’ which starts with unmatched node n in and end with a matched node m in. Now consider the path p nm under the maximum matching M. The length of p nm must be even because nodes n in and m in are both in the set V in. Therefore, the alternating path p nm must end with an unmatched node m in related to M because n in is matched by M. This contradicts the fact that n in cannot be reached by any input node related to M, which completes the proof.
The above proof implies an important fact that any of the nodes reachable from an input node can be used to replace the original input node and obtain a new MIS. Therefore, we have the following corollary:
Corollary 1: Consider an MIS D and one of its input node m ∈ D, let K m be the set of the nodes that satisfied condition 2 of Theorem 1. For every node n ∈ K m , D’ = D − {m} + {n} is an MIS.
Based on Theorem 1, the proof to the corollary is trivial. Corollary 1 suggests how to find the substituting nodes for an input node. For example, in Fig. 3, the substituting nodes of input node 4 are node 8 and node 6. If node 4 is not suitable for accepting control signals, we can use either node 8 or node 6 to substitute node 4, and the new MIS would be {8, 10} or {6, 10}.
An example of the process of algorithm All_Input(G). (A) A sample network and its maximum matching (red edges) (B) the corresponding bipartite graph; (C) In the last step of the algorithm, we search for alternating paths from unmatched nodes. The nodes on the alternating paths in V in are nodes {8, 6, 9}, and the input nodes are {4, 10}. Therefore, all possible input nodes of the network are {4, 6, 8, 9, 10}. The substituted nodes of input node 4 are node 8 and node 6, and those of input node 10 is node 9. Therefore, if node 4 or 10 is not suitable to be inputted control signals, we can use the corresponding substituted nodes to input control signals and regain full control of the network.
The significance of Theorem 1 and Corollary 1 is that all possible input nodes can be identified by some alternating paths of the input nodes of any given MIS. This observation leads to a novel two-step approach to identification of all possible input nodes, i.e., we first compute an MIS and then consider all of its alternating paths. Moreover, these two steps can be combined using a simple modification to the Hopcroft–Karp maximum matching algorithm for undirected graphs27. The basic idea of the Hopcroft–Karp algorithm is to iteratively find all augmenting paths corresponding to the matching M at hand, and then to derive a larger matching M’. A maximum matching is obtained when no augmenting path can be founded. The last step of the algorithm is exactly to look for all alternating paths starting from the input nodes of the maximum matching. Therefore, all possible input nodes can be obtained in the last step of Hopcroft–Karp algorithm based on Theorem 1.
The above idea and steps can be formulated in Algorithm All_Input(G) for finding all possible input nodes in network G, which is listed as follows:
All_Input(G):
-
1.
For a directed network G(V, E), let B(V out, V in, E) be its corresponding bipartite graph; let the initial matching M = null;
-
2.
Find all the alternating paths of all unmatched nodes in V in, denoted as AP = {P 1, P 2 … P n }, and let the nodes of AP in V in as candidate results;
-
3.
If AP contain augmenting paths, expand all augmenting paths and obtain a new matching M’; clear all candidate nodes, M = M’; return to step 2;
-
4.
If AP contain no augmenting path, the candidate nodes are all possible input nodes, and the set of the unmatched nodes is an MIS of G.
Figure 3 illustrates an example of All_Input(G) on a small network. The time complexity of the above algorithm is the same as that of the Hopcroft-Karp algorithm, which is O(N 1/2 L).
Result
To assess the efficiency of the new algorithm, which was coded in JAVA, we compared it with the previous algorithm22. The source code of our algorithm and the previous algorithm is available in the supplementary information. The comparison was done on a Windows 7 workstation with a quad-core Intel i7-3770 processor of 3.9 GHz and 32GB DDR3 1600MHz memory.
We considered 13 synthetic networks, in which the number of nodes n varied from 105 to 5 × 106 and the average degree <k> varied from 6 to 16. Networks were generated with Gephi28 based on the Scale-Free Network model29. The experimental results on these networks showed that our algorithm outputted the same results set yet significantly outperformed previous algorithm22 (Fig. 4). With a small network with n = 105, our algorithm achieved 52x speedup compared to the previous algorithm22. With a larger network with n = 5 × 106, our algorithm achieved 7330x speedup with the execution time being only 3.276 seconds. Note that the speedup increases with the average degree.<k> (Fig. 4A), which indicates that our algorithm has better performance on dense networks. The details of the results are listed in Table 1.
Next, we evaluated the performance of the algorithm on some real networks. These networks were selected based on their diversity of topological structures. These networks include biological networks, social networks, and Internet networks. The size of these networks varied from very small (E. Coli network, 423 nodes) to very large (Amazon network, 4 × 106 nodes). The results shown in Table 2 indicated that our algorithm also significantly outperformed the previous algorithm. On large networks, such as Amazon or Twitter, the results can be obtained within 10 seconds, resulting in an almost 104x speedup compared to the previous algorithm.
As we have proven in corollary 1, our algorithm can also output the substituting nodes for each input node. Figure 5A shows an example of St. Marks foodweb43, which has 13 input nodes in an MIS. We computed the substituting nodes for each input node and showed the alternating paths between them in Fig. 5B. Interestingly, the size of the substituting nodes set of each input node is different, indicating some input nodes are more robust in controlling the network. The experimental results on other real networks are similar (Fig. 6). Note that some input nodes have the same number of substituting nodes because they are linked to the same set of substituting nodes, e.g., those of TRN-Yeast-131 and Ythan foodweb44 in Fig. 6.
Input nodes and their substituted set of St. Marks foodweb43. (A) Network topology of St. Marks foodweb. The input nodes are green nodes. (B) All 13 Input nodes and their substituted nodes. We showed the alternating paths which connected the input nodes and their substituted nodes. The red edges are the matching edges.
Percentage of substituted nodes for each input node of real networks. The networks we used are Kohonen37, TRN-Yeast-131, Ythan foodweb44 and TRN-Yeast-231. The vertical axis represents the percentage of substituted nodes p i = s i /N, where s i is the number of substituted nodes of input node i, and N is the total number of the nodes. The horizontal axis shows an MIS, in which the input nodes are sorted based on s i by descending order.
Conclusion
We developed an efficient algorithm for finding all possible input nodes for controlling complex networks. We proved that all possible input nodes can be efficiently identified along with computing an MIS without increasing the overall complexity beyond finding the MIS. Therefore, our algorithm offers a significant speedup over the previous algorithm on both synthetic networks and many large real networks. Furthermore, our algorithm can also output the substituted nodes set for each input node. It means that once we computed an MIS, we can immediate obtain all the substituting nodes for the MIS. Thanks to its efficiency, the new algorithm makes it possible to study controllability of large real-world networks and will have many potential applications in diverse areas.
Data availability
All data generated or analyzed during this study are included in this article (and its Supplementary Information files).
References
Kalman, R. E. Mathematical description of linear dynamical systems. Journal of the Society for Industrial and Applied Mathematics, Series A: Control 1, 152–192 (1963).
Luenberger, D. Introduction to dynamic systems: theory, models, and applications. Proceedings of the IEEE 69 (1979).
Murota, K. Matrices and matroids for systems analysis. Vol. 20 (Springer Science & Business Media, 2009).
Vinayagam, A. et al. Controllability analysis of the directed human protein interaction network identifies disease genes and drug targets. Proc Natl Acad Sci USA 113, 4976–4981, doi:10.1073/pnas.1603992113 (2016).
Wuchty, S. Controllability in protein interaction networks. Proceedings of the National Academy of Sciences 111, 7156–7160 (2014).
Basler, G., Nikoloski, Z., Larhlimi, A., Barabási, A.-L. & Liu, Y.-Y. Control of fluxes in metabolic networks. Genome research 26, 956–968 (2016).
Liu, Y.-Y., Slotine, J.-J. & Barabási, A.-L. Controllability of complex networks. Nature 473, 167–173 (2011).
Zhang, X., Lv, T. & Pu, Y. Input graph: the hidden geometry in controlling complex networks. Scientific reports 6 (2016).
Ruths, J. & Ruths, D. Control profiles of complex networks. Science 343, 1373–1376 (2014).
Lin, C.-T. Structural controllability. IEEE Transactions on Automatic Control 19, 201–208 (1974).
Wuchty, S., Boltz, T. & Küçük‐McGinty, H. Links between critical proteins drive the controllability of protein interaction networks. Proteomics (2017).
Kawakami, E. et al. Network analyses based on comprehensive molecular interaction maps reveal robust control structures in yeast stress response pathways. npj Systems Biology and Applications 2, 15018 (2016).
Wang, B. et al. Diversified control paths: A significant way disease genes perturb the human regulatory network. PloS one 10, e0135491 (2015).
Ravindran, V., Sunitha, V. & Bagler, G. Identification of critical regulatory genes in cancer signaling network using controllability analysis. Physica A: Statistical Mechanics and its Applications 474, 134–143 (2017).
Zhang, X., Lv, T., Yang, X. & Zhang, B. Structural controllability of complex networks based on preferential matching. PloS one 9, e112039 (2014).
Menichetti, G., Dall’Asta, L. & Bianconi, G. Network controllability is determined by the density of low in-degree and out-degree nodes. Physical review letters 113, 078701 (2014).
Nacher, J. C. & Akutsu, T. Dominating scale-free networks with variable scaling exponent: heterogeneous networks are not difficult to control. New Journal of Physics 14, 073005 (2012).
Nacher, J. C. & Akutsu, T. Structural controllability of unidirectional bipartite networks. Scientific reports 3, 1647 (2013).
Delpini, D. et al. Evolution of controllability in interbank networks. Scientific reports 3, 1626 (2013).
Kumar, A., Vlachos, I., Aertsen, A. & Boucsein, C. Challenges of understanding brain function by selective modulation of neuronal subpopulations. Trends in neurosciences 36, 579–586 (2013).
Zdeborová, L. & Mézard, M. The number of matchings in random graphs. Journal of Statistical Mechanics: Theory and Experiment 2006, P05003 (2006).
Jia, T. et al. Emergence of bimodality in controlling complex networks. Nature communications 4 (2013).
Uno, T. Algorithms for enumerating all perfect, maximum and maximal matchings in bipartite graphs. Algorithms and Computation, 92–101 (1997).
Uno, T. In International Symposium on Algorithms and Computation. 367–379 (Springer).
Tassa, T. Finding all maximally-matchable edges in a bipartite graph. Theoretical Computer Science 423, 50–58 (2012).
Berge, C. Two theorems in graph theory. Proceedings of the National Academy of Sciences 43, 842–844 (1957).
Hopcroft, J. E. & Karp, R. M. In Switching and Automata Theory, 1971., 12th Annual Symposium on. 122–125 (IEEE).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. ICWSM 8, 361–362 (2009).
Maslov, S. & Sneppen, K. Specificity and stability in topology of protein networks. Science 296, 910–913 (2002).
Shen-Orr, S. S., Milo, R., Mangan, S. & Alon, U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature genetics 31, 64–68 (2002).
Balaji, S., Babu, M. M., Iyer, L. M., Luscombe, N. M. & Aravind, L. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. Journal of molecular biology 360, 213–227 (2006).
Milo, R. et al. Network motifs: simple building blocks of complex networks. Science 298, 824–827 (2002).
Vinayagam, A. et al. A directed protein interaction network for investigating intracellular signal transduction. Sci. Signal. 4, rs8–rs8 (2011).
Leskovec, J., Lang, K. J., Dasgupta, A. & Mahoney, M. W. Community structure in large networks: Natural cluster sizes and the absence of large well-defined clusters. Internet Mathematics 6, 29–123 (2009).
Leskovec, J., Huttenlocher, D. & Kleinberg, J. In Proceedings of the 19th international conference on World wide web. 641–650 (ACM).
De Nooy, W., Mrvar, A. & Batagelj, V. Exploratory social network analysis with Pajek. Vol. 27 (Cambridge University Press, 2011).
Handcock, M. S., Hunter, D., Butts, C. T., Goodreau, S. M. & Morris, M. statnet: An R package for the Statistical Modeling of Social Networks. Web page http://www.csde.washington.edu/statnet (2003).
Ripeanu, M., Foster, I. & Iamnitchi, A. Mapping the gnutella network: Properties of large-scale peer-to-peer systems and implications for system design. arXiv preprint cs/0209028 (2002).
Leskovec, J., Adamic, L. A. & Huberman, B. A. The dynamics of viral marketing. ACM Transactions on the Web (TWEB) 1, 5 (2007).
Leskovec, J. & Mcauley, J. J. In Advances in neural information processing systems. 539–547.
De Domenico, M., Lima, A., Mougel, P. & Musolesi, M. The anatomy of a scientific rumor. arXiv preprint arXiv 1301, 2952 (2013).
Opsahl, T. & Panzarasa, P. Clustering in weighted networks. Social networks 31, 155–163 (2009).
Baird, D., Luczkovich, J. & Christian, R. R. Assessment of spatial and temporal variability in ecosystem attributes of the St Marks National Wildlife Refuge, Apalachee Bay, Florida. Estuarine, Coastal and Shelf Science 47, 329–349 (1998).
Dunne, J. A., Williams, R. J. & Martinez, N. D. Food-web structure and network theory: the role of connectance and size. Proceedings of the National Academy of Sciences 99, 12917–12922 (2002).
Acknowledgements
This research was supported by the Fundamental Research Funds for the Central Universities of China under grand number N140404011, and the Natural Science Foundation of China under grant number 91546110, and China Scholarship Council under grant number 201606085011, and the Special Program for Applied Research on Super Computation of the NSFC-Guangdong Joint Fund (the second phase).
Author information
Authors and Affiliations
Contributions
X.-Z.Z. proved the theorem and designed algorithm. J.-F.H. wrote the code and performed the experiments. X.-Z.Z. and W.-X.Z. wrote the paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, X., Han, J. & Zhang, W. An efficient algorithm for finding all possible input nodes for controlling complex networks. Sci Rep 7, 10677 (2017). https://doi.org/10.1038/s41598-017-10744-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-10744-w
This article is cited by
-
Total network controllability analysis discovers explainable drugs for Covid-19 treatment
Biology Direct (2023)
-
Negative ion beam source as a complex system: identification of main processes and key interdependence
Rendiconti Lincei. Scienze Fisiche e Naturali (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.