Abstract
Metabolism estimates organic carbon accumulation by primary productivity and removal by respiration. In rivers it is relevant to assessing trophic status and threats to river health such as hypoxia as well as greenhouse gas fluxes. We estimated metabolism in 17 rivers of the Illinois River basin (IRB) for a total of 15,176 days, or an average of 2.5 years per site. Daily estimates of gross primary productivity (GPP), ecosystem respiration (ER), net ecosystem productivity (NEP), and the air-water gas exchange rate constant (K600) are reported, along with ancillary data such as river temperature and saturated dissolved oxygen concentration, barometric pressure, and river depth and discharge. Workflows for metabolism estimation and quality assurance are described including a new method for estimating river depth. IRB rivers are dominantly heterotrophic; however, autotrophy was common in river locations coinciding with reported harmful algal blooms (HABs) events. Metabolism of these regulated Midwestern U.S. rivers can help assess the causes and consequences of excessive algal blooms in rivers and their role in river ecological health.
Similar content being viewed by others
Background & Summary
Aquatic metabolism measures the balance between organic carbon accumulation by primary productivity of algae and other autotrophs and the rate of carbon removal by respiration of autotrophs and heterotrophs such as bacteria. River metabolism is relevant to assessing causes and consequences of eutrophication such as hypoxia, serving as an early warning indicator of changing river functions and health as well as indicating shifts in greenhouse gas emissions1,2. Here we focused on metabolism of regulated rivers in the Illinois River basin (IRB) where river algal blooms and associated toxins have been reported3,4,5,6,7. To quantify metabolism, the rate of oxygen production and consumption in the aquatic system is measured over time to estimate gross primary productivity (GPP) and ecosystem respiration (ER). GPP is a positive quantity that estimates the daily growth rate of autotrophs and ER is a negative quantity that estimates the daily rate of organic carbon loss by organism respiration including respiration of autotrophs and respiration associated with microbial decomposition of detrital organic matter. The sum of GPP and ER is the net ecosystem productivity (NEP), which estimates the daily balance between organic carbon build up and depletion in the system by primary productivity and respiration. To use the oxygen balance method to estimate metabolism it is necessary to also quantify the rate of dissolved oxygen exchange with the atmosphere, which depends on water temperature and atmospheric pressure as well as water mixing and turbulence. As methods improve to measure metabolism, the numbers of studies have substantially increased. However, most long-term estimates in flowing waters are confined to small streams and wadable rivers2.
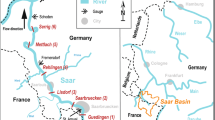
For the present study we estimated aquatic metabolism at 17 river sites in the Illinois River basin (IRB)8 that encompassed extensive agricultural areas and a major metropolitan area in northeastern Illinois as well as agricultural and suburban areas in northwestern Indiana and in southern Wisconsin that drain to the Illinois River (Fig. 1, Table 1).
The selected IRB sites represent a variety of river sizes and characteristics, including mainstem sites on the Illinois River as well as several large tributaries and a few smaller streams. The Illinois River is substantially regulated by a series of locks and dams to maintain minimum water levels for navigation through the upper Illinois River as it enters the Des Plaines River tributary and headwaters of the Chicago Area Waterway System (CAWS). Not surprisingly, water quality and ecological conditions are substantially impaired in IRB rivers, including high nutrients and suspended sediments3,4. Large tributaries of the Illinois River include the Kankakee River which drains large areas of corn and soybean agriculture and has been dredged and straightened to increase its conveyance, and now has significant problems with high turbidity and sedimentation3. The Fox River flows through agricultural areas in southern Wisconsin and then traverses the western edge of the Chicago urban corridor before joining the Illinois River5. Dam storage in the Illinois and Fox Rivers maintains significant water depths and lengthens water residence times while also increasing water clarity4. Recently, excessive plankton blooms and associated algal toxins have been observed in the Illinois and Fox Rivers5,6,7.
The type of autotrophs in water bodies (e.g., benthic vs. planktonic algae vs. submerged aquatic vegetation) depends on light availability which is affected by tree and bank shading and water-column light attenuation, disturbance frequency and severity, and other factors1,2. Benthic algae are usually thought to dominate GPP in streams and small rivers where the river bed is illuminated1,2. Many benthic algal species are adapted to shading by forest canopies, as well as the high-flow events that scour stream beds and disrupt GPP2. Planktonic algae are usually thought to dominate in lakes, reservoirs, and estuaries; however, the expectation for large rivers is less clear9. However, unshaded rivers with low or moderate turbidity have the potential for high water-column GPP from phytoplankton growth8,9.
Phytoplankton and harmful algal blooms (HABs) have increasingly been observed in large rivers and reservoirs of the Midwest and Great Plains areas of the United States such as the Kansas, Ohio, and Mississippi Rivers10,11,12,13, as well as in the Illinois River5,6,7 and elsewhere14,15. Flow extremes are moderated in regulated rivers such as the Ohio, Mississippi, and Illinois Rivers where locks and dams lengthen the water residence time and increase the water clarity in the quiescent river pools between the dams16,17. Regulated rivers also often have abundant nutrient supply3,4,5,6 which can support phytoplankton blooms during low flow periods, when water residence time is prolonged, when water is warmer than average, and when turbidity from suspended sediments is often at its lowest16,17.
Chlorophyll-a (chl-a) is often used as a measure of phytoplankton, however, riverine chl-a can reflect a myriad of algal types and is not distinctly diagnostic of phytoplankton18. Also, the relationship between chl-a and autotrophic biomass may vary greatly depending on light, nutrients, temperature, and other factors19. Use of metabolism metrics in rivers can improve understanding of the drivers of river algal blooms20 and can help anticipate future changes in river health21,22,23. For example, changes in the sign of NEP and in the temporal correlation of GPP and ER can signal changes in the relative importance of phytoplankton versus submerged aquatic vegetation as dominant primary producers in rivers21.
Most previous metabolism estimation in rivers was focused on streams and small rivers2. To motivate further use of the IRB metabolism data8, we plotted long-term average metabolism for 17 IRB river sites (Fig. 2). Like many heterotrophic streams and rivers that process substantial inputs of allochthonous organic matter1,2,9,23, the metabolism of IRB rivers was generally heterotrophic (Fig. 2).
Average gross primary productivity (GPP) versus ecosystem respiration (ER) in regulated rivers and various tributaries of the Illinois River Basin (IRB), USA. IRB study rivers are distinguished by symbol color with symbol size scaled by mean river discharge. Dashed line denotes where net ecosystem productivity (NEP) equals zero and separates heterotrophic from autotrophic conditions. The orange cross shows the approximate inter-quartile range of average GPP and ER for 18 “unshaded and stable flow” rivers in the United States2.
The overall productivity of IRB rivers (mean GPP = 2.77 g O2 m−2 d−1) was representative of the relatively high productivity of a subgroup of 18 high productivity “unshaded and stable flow” rivers evaluated as part of a study of 220 rivers and streams2 (Fig. 2). Productivity was generally higher in unshaded and stable flow rivers compared to most other streams and rivers because of greater light availability and because smaller variations of river discharge disturb autotrophs less frequently2. Only one of our IRB study rivers (Fox R. with an average GPP of 7.13 g O2 m−2 d−1) was a standout in productivity compared to the unshaded and stable flow subgroup. However, nearly all IRB rivers were substantially higher (more negative) in ER (mean ER = −6.05 g O2 m−2 d−1) compared with the unshaded and stable flow subgroup from the broader analysis2 (Fig. 2).
Our dataset indicates that IRB river metabolism is heterotrophic overall (mean IRB river NEP = −3.28 g O2 m−2 d−1), however, IRB rivers were intermittently autotrophic, accounting for between 1 and 56% of the measured days (Table 6 and Fig. 2). At one extreme the Kankakee and Des Plaines Rivers were usually strongly heterotrophic and were only autotrophic on 1% and 5% of days, respectively. At the other extreme the Illinois River and Fox Rivers were autotrophic 33% and 43% of days, respectively. Tributaries were intermediate in their autotrophy ranging between 12% and 23% of days (Table 6 and Fig. 2).
Frequent autotrophy in rivers is an indicator but does not in itself imply phytoplankton production21. However, the correspondingly high chlorophyll-a (chl-a) measurements in the Illinois and Fox Rivers6 compounded with visual reporting and analytical determinations of planktonic algae5,7 indicate that phytoplankton blooms are common in the IRB. We encourage further analysis of our IRB river metabolism data set8 in the context of water quality24,25 and river conditions26,27,28 to better understand the triggers and consequences of riverine planktonic algal blooms, in the IRB and elsewhere.
Methods
Initial site selection for metabolism estimation in IRB rivers was based on the availability of dissolved oxygen data accessed from the U.S. Geological Survey National Water Information System25 (USGS NWIS). We used the USGS | National Water Dashboard link to help identify NWIS site numbers with the needed input data. USGS scalable maps of water-quality data collection sites that are available at that site were consulted. Potential river sites were identified by searching all “stream type” sites including “streams”, “canals”, and “ditches” with at least a year of continuous collection of dissolved oxygen data (i.e., generally 15-minute intervals). Sites were excluded that were obviously not lotic in character, e.g., wetlands, ponds, gravel pits, which resulted in identifying seventeen IRB river sites that were appropriate for modeling long-term metabolism. Selected sites were linked to the National Hydrography Dataset (NHDPlus)26 to take advantage of documented river and catchment attributes.
We used USGS data retrieval software (dataRetrieval)29 to download between one and nine years of data from 17 selected IRB river sites (Table 1) including all continuous (sub-daily) measurements of dissolved oxygen concentration, water temperature, specific conductivity, continuous daily water discharge and gage height (Table 2), as well as downloading infrequently collected channel field measurements (Table 3). Barometric pressure was obtained separately through a request to NOAA30 using site latitude and longitude to select the closest nearby measurement location for each river site. All of the dissolved oxygen (DO) data used in this study were quality assured and approved by the USGS. The DO data are expected to be of high quality because they were collected after 2010, after the use of optical DO sensors had become standard practice. Although it did not apply to our IRB data, recently collected USGS data that is available for download is sometimes provisional and not yet quality assured.
To model metabolism we took advantage of recent advancements with state-space models that simultaneously estimate three unknown metabolism variables, GPP, ER, and K60031,32,33. Generally, models converge better and produce physically realistic estimates when GPP > rate of air-water oxygen exchange, a condition that accentuates diel variation in dissolved oxygen concentration and increases the signal-to-noise ratio that aids model identification of the competing influences of GPP, ER, and K600. Nevertheless, metabolism estimation remains a challenge because of the potential difficulties in estimating three co-related parameters from a single oxygen time series.
To model metabolism in IRB rivers we used the streamMetabolizer R package (https://github.com/USGS-R/streamMetabolizer), a widely tested and well documented state-space metabolism model33. This model uses the one-station modeling approach that assumes that sensor data collected at a single point in a river is representative of a well-mixed water column. The accuracy of DO measurements is also important; however, the measurement accuracy has improved substantially since high-quality optical dissolved oxygen sensors began being used routinely (approximately 2005). Furthermore, the model does not quantify anaerobic respiration that is sometimes significant in low-oxygen rivers. In addition to assuming well-mixed conditions, the one-station modeling approach assumes homogenous upstream conditions affecting metabolism for a distance that is assumed to be proportional to v/K where v is stream velocity and K is the gas exchange coefficient.
The governing mass balance equations equate the instantaneous rate of change in DO [O2] in the river with the sum of the rates of DO inputs and outputs by metabolism and gas exchange32. Expressed as volumetric rates, the mass balance for DO is:
where d[O2]/dt is the rate of change in water column O2 [mg O2 L−1 d−1]; Pt is the instantaneous volumetric rate of oxygen addition by gross primary production [mg O2 L−1 d−1]; Rt is the instantaneous volumetric rate of oxygen removal by respiration [mg O2 L−1 d−1]; and Dt is the instantaneous volumetric rate of air-water oxygen exchange [mg O2 L−1 d−1]. By the definition, Pt should be greater than or equal to zero, Rt should be less than or equal to zero, and gas exchange, Dt, can take either sign. The streamMetabolizer model33 restructured the oxygen balance expressions by using long-term oxygen times series to estimate daily metabolism variables through the solution of the following equations:
where GPP is the daily areal average rate of primary production (g O2 m−2 d−1), ER is the daily areal average rate of respiration [g O2 m−2 d−1], and K600 is the daily average gas exchange rate constant normalized for molecular properties and temperature to a Schmidt number of 600 [day−1]. Variables with subscript t are instantaneous values that are typically estimated from 15-minute interval measurements. The rate of gas exchange, Dt, is the product of the rate constant and the deficit between actual and saturated concentrations of dissolved O2. Rather than fit actual gas exchange, i.e., the K2,t value, the model fits K600, so that only one standardized gas-exchange-related parameter per day need be reported that still captures and reflects the within-day variation in gas exchange rates caused by diel variation in temperature. Additional variables are h, mean river depth representing the width and upstream length of the reach affecting the oxygen balance [m]; PPFD, photosynthetic photon flux density [μmol photons m−2 d−1]; Osat,t, saturated O2 concentration [mg O2 L−1]; Omod,t, model estimated O2 concentration [mg O2 L−1]; K2,t, O2-specific and temperature specific gas exchange coefficient [day−1]; Tt, water temperature [°C]; and S, Schmidt number coefficients: SA = 1568, SB = −86.04, SC = 2.142, and SD = −0.0216. The solution approach is described in detail in Appling et al.33.
River depth estimation
River depth is necessary for metabolism estimation and the accuracy of depth estimation has a directly proportional effect on the estimation accuracy of GPP and ER. An approach previously underutilized for depth estimation in multi-river metabolism studies is using channel field measurements by the U.S. Geological Survey. We used a linear rating curve approach for estimating river depth that was based on USGS field measurements of channel width, channel area, gage height, channel discharge and channel cross-section average velocity. We obtained those field measurements from USGS NWIS25 using the dataRetrieval29 function “readNWISmeas()” that referenced USGS NWIS site number and start and end date, which often returned tens of field measurements for each site during the period of interest.
To use the linear rating curve approach to estimate river depth, the cross-section averaged depth was determined for days with field measurements by dividing the measured flow cross section by the wetted channel width:
where hfm is the field measured river depth, Afm is the field measured channel cross-sectional area, and wfm is the field measured wetted width of the river.
River depth for all model days was estimated from a linear estimation equation:
where h and GH are river depth and measured gage height, respectively, and model coefficients m and b for this equation were determined from a linear regression of the field measured river depth against measured gage height on the days of the field measurements.
Usually, we excluded USGS field measurements rated as “poor” from the regression of field measured river depth on gage height. At some sites, however, most of the field measurements, and sometimes all of them, were rated as poor. Nevertheless, if the gaging cross section was representative of upstream conditions, we usually judged that using field measurements to estimate river depth was superior to hydraulic geometry estimation of river depth no matter what the quality rating of the field measurements. The preferred water depth estimation method for each site is noted in Table 7.
We used the linear rating curve estimation approach for estimating river depth at thirteen of the seventeen IRB river sites where the river width at the sensor location was representative of upstream conditions (see details in next section). However, four of the seventeen river sites were located at relatively narrow control sections for which river depth estimates at the sensor location were not representative of upstream conditions. For those sites we used a hydraulic geometry approach34 to estimate cross-section average river depth, h, estimated from hydraulic geometry as:
where c and f are hydraulic geometry coefficients35 for each of the river reach codes (comID26) associated with our IRB river sites, and Q is continuous discharge at the IRB river site.
Assessing site representativeness of river conditions
The one station method for estimating metabolism depends on the measurement site representing both local and upstream conditions that affect metabolism estimates. A well-mixed water column, both vertically and laterally, is assumed with longitudinal consistency in river physical and biological conditions34. Those assumptions have been examined theoretically36 but are not often tested at field sites. For the present study we assessed the consistency of river width at the oxygen sensor site with river width upstream to evaluate whether the local measured river depth was representative of upstream conditions.
It is not unusual for USGS gaging and sensor measurement cross sections to be located at “control sections” that are narrower than average for the river reach, in which case the field measurements from the cross section may differ from the reach average. Both the average river depth and average velocity could be overestimated in a narrower than average measurement cross section. We consulted the USGS “water-year summary” for each site25 and we visually examined the gaging cross section and upstream conditions using publicly available aerial imagery (https://www.google.com/maps). The sensor location and gaging cross section where depth was measured by USGS field crews was determined from the description provided in the water-year summary25. Using the imagery, we examined the consistency of river width at the measurement site for approximately 10 kilometers upstream of the oxygen measurement site. Because the regulated rivers of the IRB were relatively consistent in width, we could estimate the river depth at most sites using the linear rating curve approach as described in the previous section.
To accurately estimate river metabolism, we also had to be concerned how close the site was to upstream flow regulation structures, e.g., locks and dams, or lakes. If close enough, those features affect dissolved oxygen concentrations in ways that disrupt the river metabolic signals being modeled at the sensor site. Proximity is usually judged by estimating the “metabolism reach length”, i.e., the distance required for substantial turnover of the dissolved oxygen in the water column by gas exchange with the atmosphere. Metabolism reach length was estimated as the river distance required for 80% turnover in river dissolved oxygen by gas exchange34, i.e., the distance where upstream river conditions are likely to influence metabolism calculations. For each day in each river, we estimated the metabolism reach length as:
where v is the cross-section averaged river velocity in m d−1, and \({K}_{{O}_{2}}\) is the air-water exchange coefficient for oxygen that was calculated from the K600 using the measured water temperature and published analysis equations and coefficients33. Cross-section averaged river velocity was estimated by dividing daily average discharge by the estimated cross-sectional channel area for that day:
where Afm is the field measured channel cross-sectional area. A for each modeled day was estimated using a linear estimation equation:
where GH is gage height and m and b for this equation are model coefficients determined from a linear regression of the field measured cross-sectional channel area against measured gage height for the days of the field measurements.
To compare the estimated metabolism reach length with field conditions, we measured the distance from the metabolism sensor site to the nearest upstream flow regulation structures, e.g., lock and dam, or lake, by visual inspection of publicly available aerial imagery (https://www.google.com/maps) where we used that product’s measurement tool to estimate the distance from the metabolism sensor site.
Workflow for modeling IRB river metabolism
We used R Statistical Software37 to process existing data to create model inputs, verify model inputs, run the streamMetabolizer model, and post-process and quality assure the results (Fig. 3).
The broad outlines of the workflow are documented in Fig. 3 and Table 4 and briefly summarized here. Running the first script time-matched the downloaded data, converted units, and filled time gaps less than 3 hours by linear interpolation. Running script 2 calculated model input variables such as solar time, saturated dissolved oxygen concentration, river depth, and estimated a proxy for light intensity at the river surface, and produced an output file compatible with the requirements of streamMetabolizer. The script 2 calculations were based on published functions34, except for the new method of estimating river depth discussed in the “River depth estimation” section.
Running script 3 provided a consistency check with script 1 outputs before running script 4 to run the streamMetabolizer model. Script 5 post processes the model outputs to produce results and model diagnostics where daily metabolism results are flagged based on established criteria34. Also provided are plots for visual evaluation of the results as well as censored versions of metabolism output files that remove results for all days that were flagged. Details are provided in the “Quality assurance” section. Table 4 summarizes script operation in data acquisition, preparation of inputs, running the model, and post-processing outputs to evaluate and quality assure the model results.
Running the metabolism model
We ran streamMetabolizer version 0.12.0 on a laptop using R version 4.1.137. Computational times varied between 1 and 12 hours per site, with the two IRB sites with more than 5 years of record (Kankakee River at Davis and Illinois River at Florence) needing to be split into approximately 3-year segments to facilitate run completion. We used the streamMetabolizer option for Bayesian partial pooling in our models, which conditions estimates of K600 based on the expectation that K600 varies as a function of discharge. Appling et al.33 showed that partial pooling helps improve model performance because, although partial pooling does not impose a strict relationship between K600 and discharge, it establishes an across-day, piecewise linear relationship between ln(K600) and ln(Q) that helps improve the estimation of GPP, ER, and K600. Models were run with the recommended setup using four Monte Carlo Markov Chains and 1000 warmup steps. The streamMetabolizer model calculates values of the Gelman-Rubin statistic for observational error, \({\widehat{{\rm{R}}}}_{{{\rm{\sigma }}}_{obs}}\), process error, \({\widehat{{\rm{R}}}}_{{{\rm{\sigma }}}_{proc}}\), and K600 estimation error, \({\widehat{{\rm{R}}}}_{{{\rm{\sigma }}}_{K600}}\), with values ≤ 1.1 used as an initial screening criteria to indicate that model converged adequately38,39. Many of the IRB models converged on first run, but if unsuccessful, we ran the models again after increasing the number of burn-in steps to 1500. After the model runs were completed, we compiled the results and used the final diagnostic values reported by streamMetabolizer in our quality assurance steps. Also, at several river sites we tested the influence of using the default initial values for GPP, ER, and K600 provided in streamMetabolizer by varying initial values by approximately a factor of two and finding that model outcomes were robust.
Quality assurance
Daily model outputs were flagged based on indicators of poor signal to noise strength of the modeled timeseries, and indicators of biologically and physically unrealistic outcomes for GPP, ER, and K600. For Flag 1, we compared each day’s coefficient of determination of modeled oxygen, R2det against a threshold to assess signal to noise strength. For Flag 2 and 3, we assessed biologically unrealistic values of GPP and ER, respectively, following a previous example34 that allowed for slightly negative GPP and slightly positive ER outcomes to reflect error variation. Lastly, for Flag 4 we assessed physically unrealistic values of K600 (Table 5).
Our overall confidence assessments in metabolism outcomes followed Appling et al.34 (Table 5). We assessed the percentage of days that estimated GPP, ER, and K600 fell outside biologically or physically realistic thresholds as well as assessing model convergence statistics (\(\widehat{{\rm{R}}}\)) that could indicate inadequate convergence of parameter estimates. Lastly, we assessed potential interference in metabolism estimation depending on proximity of nearest upstream dam or lake (Table 5).
To evaluate overall confidence in metabolism results for IRB rivers, we ranked each river based on combining the individual rankings for the five criteria [(Table 5)]. A river site’s individual ratings needed to be high for all five metrics for that site’s metabolism overall output to rank as “High” in confidence. A single low rating for any criterion earned a “Low” overall confidence assessment. All other combinations of individual ratings earned a “Medium” overall confidence assessment for a river site’s estimated metabolism (Table 5).
Data Records
Our U.S. Geological Survey data release8 (https://doi.org/10.5066/P9TEBOUR) presents long-term aquatic metabolism estimation at 17 river sites in the IRB. The principal outcomes are 15,176 daily estimates of GPP, ER, and K600 accompanied by sub-daily input timeseries of dissolved oxygen, temperature, barometric pressure, and river depth and discharge, as well as diagnostic metrics and statistics which we used to assess the quality of model outcomes. Our source data for the IRB (Table 1) had only minimal overlap encompassing a partial record for one site, DES PLAINES RIVER AT JOLIET, IL, with a previous multi-river modeling study40.
Metabolism estimates for the Illinois River and Fox River indicate that autotrophic conditions occur between 14 and 56% of days compared to the Kankakee and Des Plaines Rivers, which experienced autotrophy on just a few percent of days (Table 6). Metabolism in the regulated rivers of the IRB can be informative about hydrologic, biogeochemical, and ecosystem health issues in larger rivers managed for navigation. We particularly encourage use of the IRB river metabolism data8 by joining with other IRB data sets24 to identify and isolate drivers and develop early warning indicators of planktonic algal blooms in rivers.
Data release file structure
Our data release8 provides files documenting metabolism estimation for 17 IRB rivers and the associated workflow. The main landing page of the USGS data release includes the metadata, readme file, and scripts (R code), and from there two child items that can be accessed leading to “Input data” and “Output data” pages, each with additional metadata and downloadable files. The data release can be accessed at https://doi.org/10.5066/P9TEBOUR. The structure of the data release and locations of downloadable files are summarized below:
MAIN PAGE: Metadata File, Readme File, and Scripts
-
RiverMET_workflow_and_scripts_metadata.xml: Metadata file describing overview of workflow and scripts
-
RiverMET_readMe.txt: Readme file providing overview of file contents and guidance for running the scripts
-
RiverMET_Scripts.zip: R code scripts 1 through 5 are provided and can be downloaded with this zip file. For convenience, we list the Script names and note behind each Script the input and output files that are downloadable under Child Item 1 (Inputs) and Child Item 2 (Outputs) as described further below:
-
1_Process-Data.R (note: Script-1 input files not included but output from Script-1 is provided in the form of Script-2 input files)
-
2_Prepare-Model-InputFiles.R (note: Script 2 input files included, see Child Item 1; Script-2 output files also included and are equivalent to Script-3 and Script-4 input files, see Child Item 2)
-
3_Verify-Model-InputFiles.R (note: Script-3 output files not included because this is an optional step for cross checking files)
-
4_Run-streamMetabolizer.R (note: Script-4 output files are not included because they are not useful without first being processed by Script-5)
-
5_PostProcess-ModelOutputs.R (note: Script-5 output files are included, see Child Item 2)
-
CHILD ITEM 1: Input Files
-
RiverMET_Input_Files_metadata.xml: Metadata file describing all input data including column headers and data units.
-
RiverMET_Inputs.zip: Downloadable Script 2 input files with filenames and contents summarized below.
-
barop.csv – barometric pressure in millibar (mb); 15-minute time series
-
disch_gage.csv – discharge in m3 s−1, gage height in m; 15 – minute time series
-
do.csv – dissolved oxygen in mg/L; 15-minute time series
-
sal.csv – salinity in Practical Salinity Units (PSU); 15-minute time series
-
temp.csv – water temperature in degrees Celsius (°C); 15-minute time series
-
hydraulic_coeffs.txt – hydraulic geometry coefficients a, b, c, and f as used in estimation equations for river width, B = aQb and river depth, h = cQf where Q is river discharge, B is river width, and h is river depth.
CHILD ITEM 2: Output Files
-
RiverMET_Output_Files_metadata.xml: Metadata file describing all output data including column headers and data units.
-
RiverMET_Outputs.zip: Downloadable output files in two folders, “outputs_from_script-2” and “outputs_from_script-5”. Script-2 output files are ready for modeling using streamMetabolizer. Script-5 output files are the final metabolism outputs from our study. Output files details are described below:
-
RiverMET_Outputs.zip/outputs/outputs_from script-2/: (note: 34 csv files with 17 using hydraulic geometry estimation of river depth and 17 using gage height estimation of river depth; example filename: bayesInput_[date]_depth-hgc_[site_no].csv
-
RiverMET_Outputs.zip/outputs/outputs_from_script-5/: (note: “outputs_from_script-5” has two folders, “outputs-A” and “outputs-B”. Each folder has 21 files including 15 site files plus 3 files each for 2 long-record sites. The “outputs-A” filenames follow this example: flagged_GPP_ER_K600_[date]_depth-hgc_[site_no].csv. The “outputs-B” filenames follow this example: censored_ GPP_ER_K600 _[date]_depth-hgc_[site_no].csv.
-
Technical Validation
There is no universally accepted way to quality assure modeling results. In the IRB we assessed daily metabolism results by flagging values that exceeded thresholds based on biologically or physically unrealistic values or on daily model-fit diagnostics from the streamMetabolizer model (Table 5). Overall confidence in each river site’s model outcomes was assessed using aggregated metrics and statistical diagnostics, e.g., percentages of daily values that were flagged and model convergence statistics (Table 5).
In the IRB an average of 29% of the modeled days had one or more flags. As described in the section on “Data release file structure”, two output versions were produced that can serve various needs. The first output version provides only censored GPP, ER, and K600 model estimates of the highest apparent quality after removing all days with flags. However, it is possible that some “useful” data may have been removed in the censoring process. The second output version provides complete results, including results for days with flags, which allows the user to judge each day’s data and allows users to perform custom assessments of the quality of model outcome to meet specific needs.
In terms of overall confidence in model outcomes, thirteen of the seventeen IRB river metabolism timeseries earned an overall high or medium confidence ranking (Table 7). The most frequent criterion causing a low confidence ranking was exceedance of the \({\widehat{{\rm{R}}}}_{{{\rm{\sigma }}}_{K600}}\) statistic threshold of 1.2 indicating problems with model convergence. The four river sites earning a low confidence ranking were FOX RIVER NEAR MCHENRY, IL; ILLINOIS RIVER AT FLORENCE, IL; SUGAR CREEK NEAR CHATHAM, IL; and LICK CREEK NEAR WOODSIDE, IL.
Having approximately three quarters of the IRB river sites (76%) earn a high or medium confidence ranking is only slightly lower performance than a similarly assessed set of rivers modeled by Appling et al. 34, where 84% ranked high or medium confidence. The IRB river metabolism results8 are therefore quality assured based on application of the best available diagnostic metrics and statistical criteria for models of this type. Nonetheless, it is important to consider that model confidence assessments are only guidance and do not override future investigations of model quality that may be more detailed or judged “fit for purpose”.
Usage Notes
Our data release8 provides metabolism outcomes and documents our workflow for modeling metabolism at 17 ILB river sites. Here we summarize descriptive information about the dataset and guidance for its use, including geographic coordinates and period of data availability for each site (Table 1), summary of USGS parameter codes used for downloading (Table 2), information about calculating parameters needed as model inputs (Table 3), an overview of script workflows (Table 4), quality assurance criteria (Table 5), and metabolism outcomes (Table 6) including a model performance assessment (Table 7). In addition, our data release8 provides guidance for potential reuse of codes in the file RiverMET_readMe.txt, including suggestions for changes that may be needed to run on a different system, re-run IRB sites with different options, or adapt scripts to model metabolism in other rivers. Users who wish to adapt parts of our workflow will need to acquire publicly available data from USGS and NOAA. They can use existing software (dataRetrieval29) to download the needed USGS data from their sites of interest, including dissolved oxygen, water temperature, specific conductance, discharge, gage height, and field measurements of channel parameters from the USGS NWIS site, and they can obtain barometric pressure data from NOAA. After downloading their own data, users can adapt parts of 1_Process-Data.R to perform the data time matching, gap filling, and unit conversion (Table 4). As long as their code produces output files that match the input files for 2_Prepare-Model-InputFiles.R that we provide in our data release, they can likely make minor adaptations to run scripts 2, 3, 4 and 5 (as described in Table 4) to prepare final model inputs, run streamMetabolizer, and organize and quality assure their metabolism modeling results.
Our data release8 also suggests approaches that can help expand the capacity for modeling river metabolism. For example, several of the IRB sites could perhaps have been included in an earlier study40 , however, not all the needed input data were available at certain sites, resulting in those sites being passed over. To facilitate modeling at those sites, where appropriate, we acquired the missing measurements from nearby “replacement” sites (Table 7). An example is several sites where dissolved oxygen was collected without collecting the river discharge needed to accomplish Bayesian partial pooling that estimates K600 based on a prior expectation that K600 varies as a function of discharge. In such cases we “replaced” the missing discharge with data from a nearby site, which allowed metabolism estimation at sites previously overlooked because of missing data8. Because of the large river size where replacement discharges were used, e.g., often over 350 m3 s−1, and given the proximity of the replacement site, usually within 10-km, we did not perform scaling by basin size when applying a replacement discharge.
Code availability
Our workflow includes scripts that were written and tested using R version 4.1.1. The scripts can be accessed from the data product8 which includes an appropriate licence (CC0 1.0 Universal) license permitting reuse without restrictions.
References
Battin, T. J. et al. River ecosystem metabolism and carbon biogeochemistry in a changing world. Nature 613, 449–459, https://doi.org/10.1038/s41586-022-05500-8 (2023).
Bernhardt, E. S. et al. Light and flow regimes regulate the metabolism of rivers. Proceedings of the National Academy of Science 119(8), e2121976119, https://doi.org/10.1073/pnas.2121976119 (2022).
McIsaac, G. F., Hodson, T. O., Markus, M., Bhattarai, R. & Kim, D. C. Spatial and Temporal Variations in Phosphorus Loads in the Illinois River Basin, Illinois USA. J Am Water Resour Assoc. https://doi.org/10.1111/1752-1688.13054 (2023).
Houser, J.N., ed. Ecological status and trends of the Upper Mississippi and Illinois Rivers (ver. 1.1, July 2022): U.S. Geological Survey Open-File Report 2022–1039, 199 p. https://doi.org/10.3133/ofr20221039 (2022).
Illinois Environmental Protection Agency, Illinois Officials Confirm Algal Bloom on Portions of the Illinois River, Residents should continue to use caution when recreating and be aware of blue-green algae. News Release June 25, 2018, Illinois Department of Public Health (2018).
Getahun, E., Keefer, L., Chandrasekaran, S. & Zavelle, A. Water Quality Trend Analysis for the Fox River Watershed: Stratton Dam to the Illinois River. Illinois State Water Survey Prairie Research Institute, University of Illinois at Urbana-Champaign prepared for the Fox River Study Group (2019).
Fox River Study Group. Fox River Implementation Plan, A plan to improve dissolved oxygen and reduce nuisance algae in the Fox River, https://www.foxriverstudygroup.org/ (2015).
Choi, J., Quion, K. M., Reed, A. P. & Harvey, J. W. RiverMET: Workflow and scripts for river metabolism estimation including Illinois River Basin application, 2005 - 2020. U.S. Geological Survey data release https://doi.org/10.5066/P9TEBOUR (2022).
Hoellein, T.J., Bruesewitz, D.A., & Richardson, D.C., Revisiting Odum (1956): A synthesis of aquatic ecosystem metabolism. Limnology and Oceanography, 58(2013), https://doi.org/10.4319/lo.2013.58.6.2089 (1956).
Manier, J. T., Haro, R. J., Houser, J. N. & Strauss, E. A. Spatial and temporal dynamics of phytoplankton assemblages in the upper Mississippi River. River Research and Applications 37(10), 1451–1462, https://doi.org/10.1002/rra.3852 (2021).
Giblin, S. M. & Gerrish, G. A. Environmental factors controlling phytoplankton dynamics in a large floodplain river with emphasis on cyanobacteria. River Res Applic. 36, 1137–1150, https://doi.org/10.1002/rra.3658 (2020).
Graham, J.L., Ziegler, A.C., Loving, B.L., & Loftin, K.A. Fate and transport of cyanobacteria and associated toxins and taste-and-odor compounds from upstream reservoir releases in the Kansas River, Kansas, September and October 2011: U.S. Geological Survey Scientific Investigations Report 2012–5129, 65 p. (Revised November 2012), https://doi.org/10.3133/sir20125129 (2012).
Graham, J. L. et al. Cyanotoxin occurrence in large rivers of the United States. Inland Waters 10(1), 109–117, https://doi.org/10.1080/20442041.2019.1700749 (2020).
Rousso, B. Z., Bertone, E., Stewart, R. & Hamilton, D. P. A systematic literature review of forecasting and predictive models for cyanobacteria blooms in freshwater lakes. Water Research 182, 115959, https://doi.org/10.1016/j.watres.2020.115959 (2020).
Beaver, J. R., Tausz, C. E., Scotese, K. C., Pollard, A. I. & Mitchell, R. M. Environmental factors influencing the quantitative distribution of microcystin and common potentially toxigenic cyanobacteria in U.S. lakes and reservoirs. Harmful Algae 78, 118–128, https://doi.org/10.1016/j.hal.2018.08.004 (2018).
Nietch, C. T. et al. Development of a Risk Characterization Tool for Harmful Cyanobacteria Blooms on the Ohio River. Water 14, 644, https://doi.org/10.3390/w14040644 (2022).
Houser, J. N., Bartsch, L. A., Richardson, W. B., Rogala, J. T. & Sullivan, J. F. Ecosystem metabolism and nutrient dynamics in the main channel and backwaters of the Upper Mississippi River. Freshw Biol 60, 1863–1879, https://doi.org/10.1111/fwb.12617 (2015).
Peipoch, M. & Ensign, S. H. (2022), Deciphering the origin of riverine phytoplankton using in situ chlorophyll sensors. Limnol. Oceanogr. Lett 7, 159–166, https://doi.org/10.1002/lol2.10240 (2022).
Cloern, J. E., Grenz, C. & Vidergar-Lucas, L. An empirical model of the phytoplankton chlorophyll: carbon ratio-the conversion factor between productivity and growth rate. Limnol. Oceanogr. 40, (1995).
Reisinger, A. J. et al. Water column contributions to the metabolism and nutrient dynamics of mid-sized rivers. Biogeochemistry 153, 67–84, https://doi.org/10.1007/s10533-021-00768-w (2021).
Diamond, J. S. et al. Metabolic regime shifts and ecosystem state changes are decoupled in a large river. Limnol Oceanogr 67, S54–S70, https://doi.org/10.1002/lno.11789 (2022).
Batt, R. D., Carpenter, S. R., Cole, J. J., Pace, M. L. & Johnson, R. A. Changes in ecosystem resilience detected in automated measures of ecosystem metabolism during a whole-lake manipulation. Proc. Natl. Acad. Sci. 110, 17398–17403 (2013).
Hall, R. O. Jr. Metabolism of streams and rivers: Estimation, controls, and application (chapter 4. In Jones, J.B. & Stanley, E.H. eds. Stream Ecosystems in a Changing Environment, 151-173 (Academic Press, 2016).
Platt, L. R. C. et al. Harmonized discrete and continuous water quality data in support of modeling harmful algal blooms in the Illinois River Basin, 2005 - 2020. U.S. Geological Survey data release https://doi.org/10.5066/P9RISQGE (2022).
U.S. Geological Survey. National Water Information System (USGS Water Data for the Nation), accessed November 4, 2021, at http://waterdata.usgs.gov/nwis/ (2021).
Blodgett, D., & Johnson, M. nhdplusTools: Accessing and Working with the NHDPlus (Version 0.5.7). Reston, VA: U.S. Geological Survey https://doi.org/10.5066/P97AS8JD (2022).
Schwarz, G. E. E2NHDPlusV2_us: Database of Ancillary Hydrologic Attributes and Modified Routing for NHDPlus Version 2.1 Flowlines. U.S. Geological Survey data release https://doi.org/10.5066/P986KZEM (2019).
Wieczorek, M. E., Jackson, S. E. & Schwarz, G. E. Select Attributes for NHDPlus Version 2.1 Reach Catchments and Modified Network Routed Upstream Watersheds for the Conterminous United States [Data set]. U.S. Geological Survey. https://doi.org/10.5066/F7765D7V (2018).
De Cicco, L.A., Hirsch, R.M., Lorenz, D., Watkins, W.D., Johnson, M. dataRetrieval: R packages for discovering and retrieving water data available from Federal hydrologic web services, v.2.7.13, https://doi.org/10.5066/P9X4L3GE (2023).
National Oceanic and Atmospheric Administration National Centers for Environmental Information. U.S. Local Climatological Data (LCD), accessed September 7, 2021, at https://www.ncei.noaa.gov/maps/lcd/ (2021).
Holtgrieve, G. W., Schindler, D. E., Branch, T. A. & A’mar, Z. T. Simultaneous quantification of aquatic ecosystem metabolism and reaeration using a Bayesian statistical model of oxygen dynamics. Limnology and Oceanography 55(3), 1047–1062, https://doi.org/10.4319/lo.2010.55.3.1047 (2010).
Grace, M. R. et al. Fast processing of diel oxygen curves: Estimating stream metabolism with BASE (BAyesian Single-station Estimation). Limnology and Oceanography: Methods 13(3), 103–114, https://doi.org/10.1002/lom3.10011 (2015).
Appling, A. P., Hall, R. O. J., Yackulic, C. B. & Arroita, M. Overcoming equifinality: Leveraging long time series for stream metabolism estimation. Journal of Geophysical Research: Biogeosciences 123, 624–645, https://doi.org/10.1002/2017JG004140 (2018).
Appling, A. P. et al. The metabolic regimes of 356 rivers in the United States. Sci. Data. 5, 180292, https://doi.org/10.1038/sdata.2018.292 (2018).
Gomez-Velez, J. D., Harvey, J. W., Cardenas, M. B. & Kiel, B. Denitrification in the Mississippi River network controlled by flow through river bedforms. Nature Geoscience 8, 941–945, https://doi.org/10.1038/ngeo2567 (2015).
Chapra, S. & Runkel, R. Modeling impact of storage zones on stream dissolved oxygen. Journal of Environmental Engineering, Volume 125, Issue 5, https://doi.org/10.1061/(ASCE)0733-9372(1999)125:5(415) (1999).
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing https://www.R-project.org (2022).
Brooks, S. P. & Gelman, A. General methods for monitoring convergence of iterative simulations. Journal of Computational and Graphical Statistics 7(4), 434–455, https://doi.org/10.1080/10618600.1998.10474787 (1998).
Gelman, A. & Rubin, D. B. Inference from iterative simulation using multiple sequences. Statistical Science 7(4), 457–472, https://doi.org/10.1214/ss/1177011136 (1992).
Appling, A. P. et al. Metabolism estimates for 356 U.S. rivers (2007–2017). U.S. Geological Survey data release https://doi.org/10.5066/F70864KX (2018).
Acknowledgements
This work was completed as part of the USGS Proxies Project, an effort supported by the USGS Water Mission Area (WMA) Water Quality Processes program to develop estimation methods for harmful algal blooms (HABs), per- and polyfluoroalkyl substances (PFAS), and metals, at multiple spatial and temporal scales. Sincere thanks are due to many who supported this effort. We extend our appreciation to Ariel Reed for her analysis contributions in the early phases of the study, Lindsay Platt for her assistance with data acquisition, Mike Stouder for preparing the metadata, Elizabeth Nystrom for reviewing the data and metadata, and Katie Summers and Jacob Zwart for their helpful reviews of an earlier draft of this manuscript. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Author information
Authors and Affiliations
Contributions
J.W.H. conceived the project and led study design, site selection, quality assurance, and writing of the manuscript. J.C. led the computations, assembled the final data release, and contributed to writing the manuscript. K.Q. made computations and contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Harvey, J.W., Choi, J. & Quion, K. Metabolism Regimes in Regulated Rivers of the Illinois River Basin, USA. Sci Data 11, 211 (2024). https://doi.org/10.1038/s41597-024-03037-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03037-1