Abstract
Extracellular vesicles play major roles in cell-to-cell communication and are excellent biomarker candidates. However, studying plasma extracellular vesicles is challenging due to contaminants. Here, we performed a proteomics meta-analysis of public data to refine the plasma EV composition by separating EV proteins and contaminants into different clusters. We obtained two clusters with a total of 1717 proteins that were depleted of known contaminants and enriched in EV markers with independently validated 71% true-positive. These clusters had 133 clusters of differentiation (CD) antigens and were enriched with proteins from cell-to-cell communication and signaling. We compared our data with the proteins deposited in PeptideAtlas, making our refined EV protein list a resource for mechanistic and biomarker studies. As a use case example for this resource, we validated the type 1 diabetes biomarker proplatelet basic protein in EVs and showed that it regulates apoptosis of β cells and macrophages, two key players in the disease development. Our approach provides a refinement of the EV composition and a resource for the scientific community.
Similar content being viewed by others
Introduction
Extracellular vesicles (EVs) are membrane bilayer-bound particles containing lipids, proteins, nucleic acids, and saccharides that are secreted by cells1. EVs are mainly classified as exosomes or ectosomes depending on their biogenesis2. Exosomes range from 30 to 200 nm in diameter and are formed via the endocytic pathway, leading to the formation of multivesicular bodies, which are then fused to the plasma membrane and released as EVs2. Ectosomes represent a variety of EV types, including microvesicles and apoptotic bodies, that buds directly from the plasma membrane2. Microvesicles are EVs of 100 to 1000 nm in diameter, whereas apoptotic bodies are larger EVs (>1000 nm) that are formed by blebbing of cells undergoing apoptosis1,2,3,4. EVs have immense potential as biomarkers, as they can carry signatures of the tissues of origin and processes affected by disease4. However, a main challenge to studying EV function and its potential as disease biomarkers is obtaining pure preparations of EVs from biofluids. This is due to the co-presence of high amounts of contaminants such as lipoproteins and albumin5,6, which share some physicochemical properties7,8.
Several analytical techniques have been developed for the isolation of EVs, including ultracentrifugation (UC), density gradient ultracentrifugation (DGUC), cushion ultracentrifugation (CUC), polymer-based precipitation (PP), size-exclusion chromatography (SEC) and immunocapture (IC)7,8. These techniques have individual advantages but also suffer from unique contaminant profiles7,8. For instance, UC co-precipitates particles of similar density, while SEC co-fractionates particles of similar sizes7,8. To improve the rigor of the EV preparation protocols, the International Society for Extracellular Vesicles (ISEV) developed a guideline with recommendations on experimental design and reporting results9, but obtaining pure EV preparation with high yields is still a challenge. Sequential purification steps have also been explored, but they can result in substantial sample losses, with EV recovery as low as 1% after two purification steps10,11. Furthermore, their labor intensiveness makes application to clinical biomarker studies challenging, given the need for large numbers of samples for adequate statistical power. Therefore, there is still a need to systematically evaluate different EV isolation techniques to better understand their performance and the nature of their contaminants.
Our team has been interested in understanding the roles of EVs in type 1 diabetes (T1D) development and their potential as biomarkers of the disease, as we discussed in a recent review3. It has been shown that EVs carry microRNAs and chemokines that can induce β-cell apoptosis12,13,14. However, more systematic studies are needed to refine the composition of EVs to facilitate the testing of individual components on T1D development.
Re-use of data deposited in public repositories allows for combined comprehensive analysis to be performed that otherwise would not be possible with data collected in a single study. This type of study is called meta-analysis and provides an opportunity to answer new questions or re-affirm/refute conclusions from previous studies15. In meta-analyses, studies are systematically searched in the literature and included or excluded with well-defined criteria, allowing for a combined analysis of the data from the different studies16. Here, we performed a meta-analysis of published proteomics data to refine the protein composition of plasma EVs. Considering that purification procedures have different performances, resulting in distinct EV/contaminant ratios6, we took advantage of these ratio differences to cluster EV-specific proteins separately from contaminants based on protein abundance profiles. This resulted in clusters enriched in EV markers, therefore, highly likely to be bona fide EV proteins, which were separated from well-known contaminant proteins. We performed a systematic review of the literature to validate the enrichment of EV proteins in these clusters. As a test case, we validated one of the proteins from the EV-enriched clusters as a bona fide EV protein using an imaging method, which is not prone to false-positive results due to contaminants in the sample. We also compared the clusters enriched with EV proteins to study their functions and their potential as biomarker candidates for T1D. We demonstrate that the meta-analysis is a powerful approach to refine EV composition and provide a better understanding of its biological roles. In addition, the refined list of plasma EV proteins represents a resource for future mechanistic and biomarker studies.
Materials and Methods
Datasets
Mass spectrometry proteomics datasets of human plasma EVs were searched in PRIDE17 and MassIVE18, the main data repositories for untargeted proteomics of the ProteomeXchange consortium. The searches were done in November and December 2021, using the keywords “extracellular vesicles”, “exosomes”, “microvesicles”, “plasma” and “serum”, following the PRISMA guidelines for meta-analysis16. Only label-free proteomics data collected on Thermo Orbitrap instruments with data-dependent acquisition were used (Fig. 1) to allow a consistent data analysis workflow. Isobaric tag labeling and low-resolution ion trap data would not allow to quantify the data intensity-based absolute quantification (iBAQ) method19, which we used for quantification. To reduce potential variability due to pathogenesis processes, only data from extracellular vesicles isolated from the plasma of healthy humans were used in our analysis. Study inclusion and exclusion criteria are listed in Fig. S1. The datasets, along with their ProteomeXchange identifiers, publications, and relevant details of the experimental methods, are listed in Table 1.
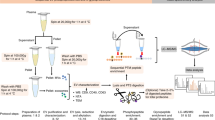
Proteomics meta-analysis of plasma extracellular vesicles (EVs). (a) Approaches: proteomics data from plasma EVs purified with a variety of methods were downloaded from ProteomeXchange, processed with MaxQuant and submitted to clustering analysis. Abbreviations: C - centrifugation, CUC - cushion ultracentrifugation, DGUC - density gradient ultracentrifugation, DUC - dilution followed by ultracentrifugation, PP - polymer-based precipitation, PROSPR - PRotein Organic Solvent PRecipitation, SEC - size-exclusion chromatography, UC - ultracentrifugation. (b) Highest enriched cluster with the top 100 extracellular vesicle proteins from Vesiclepedia when testing different numbers of clusters. P-values were calculated by Fisher’s exact test based on the distribution of expected vs. detected proteins from the top 100 Vesiclepedia proteins in each cluster. (c) Classical EV markers found in clusters 10 and 11.
Data processing
Data were processed with MaxQuant software (v.1.6.14)20 by matching against the human reference proteome database from UniProt Knowledgebase (downloaded on November 27, 2021). Searching parameters included protein N-terminal acetylation and oxidation of methionine as variable modifications, and carbamidomethylation of cysteine residues as fixed modification when appropriate. Mass shift tolerance was used as the default setting of the software: 20 ppm for the first search and 4.5 ppm for the second round. Only fully tryptic-digested peptides were considered, allowing up to two missed cleaved sites per peptide. Each data set was filtered with a 1% false-discovery rate at both peptide-spectrum and protein levels, resulting in a 2% false-discovery rate when all datasets were combined. Quantification of proteins was done using intensity-based absolute quantification iBAQ values extracted with MaxQuant. These values were further normalized by the total intensity of the whole sample to calculate the relative copy number of each protein for comparing proteins across different studies. The relative copy numbers were used to compare protein abundances across the different sets of data.
Clustering and enrichment
Clustering analysis was performed using the protein abundances with MultiExperiment Viewer – MeV (v. 4.9.0)21. Missing values were imputed with 1/10 of the smallest value in the whole dataset to set a background level, which prevents the software to overfit the data. Clustering was done by using the k-means clustering method with Pearson correlation and a maximum of 50 iterations across all individual datasets. The k-means method was used because it separates proteins into distinct clusters. To determine the optimal number of clusters, we ran separate analyses with 10, 15, 20, and 25 clusters. The optimal number of clusters was determined by cross-checking the enrichment of the top 100 proteins that appear the most often in the literature and have been made into a resource in Vesiclepedia22. Enrichment was calculated based on fold-enrichment and statistical significance with Fisher’s exact test. Principal Component Analysis (PCA) was employed after eliminating proteins with missing values, resulting in the use of 51 proteins for this analysis. The goal of PCA was to reduce the data’s dimensionality and visualize the variance in the methods used for EV extraction and identify patterns and relationships among proteins. A Python script was developed to create a PCA graph, demonstrating the distribution of samples in a lower-dimensional space, and it is available on GitHub23 (Fig. S2).
Systematic review of validated human EV proteins
The literature searches were done in PubMed on June 28, 2023, following the PRISMA guidelines for systematic reviews16. The used keywords were “extracellular vesicles”, “EV”, “microvesicles”, “MV”, “exosome” AND “immunogold” AND “human”. Studies that had no associated full text, were not conducted in human samples, were not conducted on EVs, that did not target proteins or did not use immunogold electron microscopy were excluded from the final list. The inclusion and exclusion criteria are listed in Fig. S3. Enrichment was calculated based on fold-enrichment and statistical significance with Fisher’s exact test, while the true positive rate was calculated with the following formula:
True-positive rate = (true positives)/(true positives + false negatives), being true positives the validated EV proteins that were found in EV-enriched clusters and false negatives, the validated EV proteins that were found in other clusters.
Functional- and cell-enrichment analysis
Functional-, tissue- and cell-enrichment analyses were done with DAVID24 using the KEGG database with default parameters. Specific tissue markers were mapped by comparing the data against a previously published human tissue proteomics dataset25. Specific pathways were curated with Vanted26. A script was written in R to plot a bubble graph, and it is available on GitHub23. Networks were built and plotted with Cytoscape (v. 3.9.1)27. Pathway analysis was also conducted with Ingenuity Pathway Analysis (Qiagen).
Cell growth and apoptosis assay
Murine MIN6 β cell and Raw 264.7 macrophage cell lines were cultured in DMEM containing 10% FBS and 1% penicillin-streptomycin and maintained at 37 °C in a 5% CO2 atmosphere. Cells were seeded one day prior to being treated with varying concentrations of recombinant platelet basic protein (R&D, catalog number 1091-CK-025/CF) for 24 h followed by an additional 24 h cytokine cocktail (100 ng/mL IFN-γ: R&D, catalog number 485-MI-100, 10 ng/mL TNF-α: R&D, catalog number 410-MT-010, and 5 ng/mL IL-1β: R&D, catalog number 401-ML-005) treatment. Apoptosis was measured by caspase-Glo 3/7 assay (Promega, catalog number G8092), according to the manufacturer’s instructions.
Exosome isolation and detection from human plasma
Deidentified human plasma was purchased from BioIVT. To avoid the components contained in plasma from interfering with the detection of exosomes, the plasma was purified by size exclusion chromatography (70 nm qEVsingle, IZON) to separate plasma components from exosomes. The fractions containing exosomes were incubated overnight on ExoView® chips (NanoView Biosciences) pre-coated with capture antibodies against human CD9, CD63, CD81, and negative control IgG. To detect proteins carried by exosomes, the chips were further incubated with fluorescently (CF488A)-labeled anti-CXCL7 antibody (Biorbyt, orb667781). The processed chips were scanned in the ExoView® instrument (NanoView Biosciences), and the results were analyzed with the NanoView analysis software.
Results
Meta-analysis and data processing
Plasma EV proteomics datasets were searched in the main public repositories for untargeted proteomics associated with the ProteomeXchange consortium, MassIVE, and PRIDE, identifying 24 potential studies. To keep the datasets consistent and allow to perform a single quantification method, only label-free data collected by data-dependent acquisition in orbitrap mass spectrometers were used in the study. This allowed us to use the intensity-based absolute quantification (iBAQ) method and normalize the datasets by relative copy number, enabling the comparison across different experiments. We also removed studies that were from the analysis of post-translational modifications (phosphorylation and glycosylation), that had no raw data files, insufficient metadata details and no biological replicates. One additional study was removed based on the low coverage. A total of 7 studies were eligible for the meta-analysis (see flow diagram for inclusion/exclusion criteria Fig. S1). To further make the meta-analysis consistent, we excluded samples associated with disease, and only the 105 mass spectrometry datasets from control samples were used (see characteristics in Table 1 with number of samples and purification method) (Fig. 1a). The data were processed with MaxQuant for peptide/protein identification and quantification, leading to the identification of 4,611 proteins (Table S1). A principal component analysis showed that the samples cluster based on the separation method (Fig. S2), supporting the idea that each method provides a different profile. We performed clustering analysis to group proteins based on their abundance distribution. To have discrete groups, proteins were clustered by the k-means method. The ideal number of clusters were tested in increments of 5 from 10 to 25. We evaluated the ideal number of clusters by performing an enrichment analysis of the top 100 EV proteins from the Vesiclepedia database (Table S1) and considered the ideal number of clusters to be the ones that provided a cluster with the best enrichment of the top 100 EV proteins. A total of 15 clusters were found to exhibit the best enrichment of the top 100 EV proteins (Fig. 1b, Table 2). Among the 15 clusters, clusters 10 and 11 were significantly enriched with the top 100 EV proteins from Vesiclepedia (1.3- and 1.7-fold, respectively; p-values of 0.02583 and 0.00139, respectively).
As the Vesiclepedia top 100 proteins were selected based on how often they appear in the literature, it is likely that contaminants that are commonly associated with EVs are included in that list22. Therefore, we performed a systematic review of the literature to obtain an improved list of human EV proteins that were validated by immunogold electron microscopy, a gold-standard technique to validate the localization of proteins in EVs. The literature search resulted in 204 papers that we examined manually and 48 matched our inclusion criteria, as shown in Fig. S3. A total of 47 EV proteins validated by immunogold electron microscopy were identified (Table S2). From this list, 28 identified were also found in our proteomics meta-analysis, of which 20 were grouped into clusters 10 and 11 (Fig. 1c), representing a 71.4% true-positive rate with a significant enrichment of 1.9-fold (Fisher’s exact test p-value = 0.00018917). Overall, the clustering analysis was effective in enriching for EV proteins.
Contaminants
We examined the profile of well-known contaminants (human serum albumin, lipoproteins - based on the apolipoprotein subunits) of plasma EV preparations to better understand the performance of different methods. For methods based on centrifugation (centrifugation - C, cushion ultracentrifugation - CUC, dilution followed by ultracentrifugation - DUC and ultracentrifugation - UC), 10% to 35% of the total proteome consisted of proteins were human serum albumin and lipoproteins (Fig. 2a). Compared to the UC, the additional step of CUC failed to reduce the proportion of contaminants in the samples, or to increase the proportion of EV proteins in clusters 10 and 11 (Fig. 2b). The best recovery of proteins from cluster 11 was achieved with C only (Fig. 2b). Protein Organic Solvent PRecipitation (PROSPR) had a similar contamination profile compared to the centrifugation, but with lower recovery of clusters 10 and 11. Polyethylene glycol polymer precipitation (PP) followed by density gradient ultracentrifugation (DGUC) highly enriched for lipoproteins, with apolipoprotein subunits accounting for over 50% of the total protein abundance in the sample (Fig. 2a). When PP was followed by size-exclusion chromatography (SEC), a similar proportion of contaminants was observed, but the serum albumin was the major contaminant (Fig. 2a). SEC alone had approximately 35% of the sample comprised of the 3 analyzed contaminants, predominantly lipoproteins (Fig. 2a). However, SEC alone yielded the best recovery of cluster 10 and the second highest for cluster 11, with these 2 clusters representing approximately 35% of the sample proteome combined (Fig. 2b).
Relative abundance of proteins across different extracellular vesicle purification methods and clusters of the proteome meta-analysis. The proteome samples were quantified by intensity-based absolute quantification (iBAQ) and each protein was normalized to the total sample amount. (a) Abundances of common extracellular vesicle preparation contaminants across different purification methods. (b) Abundances across different purification methods of clusters 10 and 11, which are enriched in EV markers. (c) Abundances of common extracellular vesicle preparation contaminants across different clusters of the proteome meta-analysis. Abbreviations: C - centrifugation, CUC - cushion ultracentrifugation, DGUC - density gradient ultracentrifugation, DUC - dilution followed by ultracentrifugation, PP - polymer-based precipitation, PROSPR - PRotein Organic Solvent PRecipitation, SEC - size-exclusion chromatography, UC - ultracentrifugation.
We examined the profile of serum albumin and lipoproteins across 15 clusters. Clusters 10 and 11, which showed the best enrichment in EV proteins, had a low amount of these contaminants in contrast to cluster 1 that showed a high amount of albumin, over 15% (Fig. 2c). Clusters 4, 13, and 15 were abundant in apolipoproteins, ranging from 1.5% to 6.5% (Fig. 2c). These results showed that clusters 10 and 11 were enriched in EV proteins and depleted of common plasma contaminants.
Markers of plasma EVs
To study cellular markers present in plasma EVs, we performed a cell-enrichment analysis using Database for Annotation, Visualization, and Integrated Discovery (DAVID). The analysis was based on the UniProt Knowledgebase tissue database. It showed that clusters 10 and 11 were overrepresented in proteins from blood and immune cells, including platelets, erythrocytes, T cells, B cells, monocytes, and dendritic cells (Fig. 3a). The clusters were also enriched in proteins from fibroblasts, keratinocytes, adipocytes, and Cajal-Retzius cells, a type of cortical neurons (Fig. 3a). To look for proteins that can be used as markers of specific cells, we examined the cluster of differentiation (CD) antigens, which are cell surface proteins used to distinguish cell populations by immunoassays. We found that CD antigens were enriched in cluster 10 while depleted in the other clusters. Out of 188 total CD antigens identified, 133 cell-specific CD antigens were present in cluster 10 (Fig. 3b,c, Table S3). These included markers of specific cells, such as CD45 (leukocyte, PBMC), CD3 (T cell), CD4 (T-helper cell), CD8 (cytotoxic T cell), CD19, CD20 and CD34 (B cell), CD14 and CD11b (monocyte), CD47 (reticulocyte), CD16a and CD16b (granulocyte), CD177 and CD312 (neutrophil), CD36 and CD235a (erythrocyte), CD41, CD42a, CD42b and CD16 (platelet), and CD146 (endothelial cells) (Fig. 3a). These results show that the plasma EVs carries dozens of CD markers that can be further used for immunology and cell biology experiments.
Cellular markers. (a) Cells enriched in proteins from clusters 10 and 11 in the DAVID analysis. Node (circle) sizes represents number of proteins and colors, the fold enrichment. (b) Number of CD antigens and enrichment (compared to the total identified proteins) across different clusters. Significance was determined by Fisher’s exact test. (c) Network of CD antigens across different clusters. Classical extracellular vesicle markers are colored in orange.
Pathways enriched in EV proteins
To investigate possible functions of plasma EVs, we performed a functional-enrichment analysis using the DAVID tool and the KEGG annotation (the complete list of enriched pathways is available in Open Science Framework28). Clusters 10 and 11 were enriched in proteins from several cell-to-cell communication pathways, such as pathways on antigen processing and presentation, ECM-receptor interaction, PD-L1 expression and PD-1 checkpoint, B-cell receptor signaling, chemokine signaling, and T-cell receptor signaling (Fig. 4). These clusters were also enriched in proteins from other cell signaling pathways, such as apoptosis, calcium signaling, Fc γ receptor-mediated phagocytosis, insulin signaling, insulin secretion pathway, and NOD-like receptor signaling pathways (Fig. 4). Clusters 10 and 11 were also overrepresented in lipid (estrogen, phosphatidylinositol, phospholipase D, and sphingolipid)-mediated signaling pathways (Fig. 4). Metabolic proteins, such as from the citrate cycle, cysteine and methionine metabolism, fatty acid metabolism, glycolysis/gluconeogenesis, and pentose phosphate pathways, were enriched, especially in cluster 11 (Fig. 4). Proteins from DNA replication, RNA transcription (RNA polymerase and spliceosome), and protein translation (ribosome and aminoacyl-tRNA biosynthesis) processes were enriched in cluster 9 (Fig. 4), which was not enriched with EV markers (Table 2). These results suggest that EVs may play a role in diverse communication and signaling pathways in addition to being enriched with specific metabolic pathways.
Plasma EV proteins as potential type 1 diabetes biomarkers
We next evaluated the potential of plasma EV proteins as biomarker candidates. For this, we compared our data with the human plasma proteins deposited in PeptideAtlas (4608 proteins). PeptideAtlas is an invaluable resource containing not only peptides from proteins identified in plasma, but also information on peptides with established assays for targeted proteomics, a main technique used for biomarker analyses29. By matching against the 1717 proteins from clusters 10 and 11 with the PeptideAtlas, we found 1069 common proteins (Fig. 5a, Table S4). Therefore, selecting targets among these 1069 proteins yields a higher chance of generating successful targeted proteomics assays. To further explore the potential of these proteins as biomarkers, we compared the 1069 proteins with a list of 266 T1D biomarker candidates from a systematic review that we recently conducted30. This analysis resulted in a list of 41 plasma EV proteins that are also T1D biomarker candidates (Fig. 5a, Table S5). As changes in proteins that make them biomarkers are often drivers or consequences of the pathological process of the diseases, we took a closer look at the 41 plasma EV proteins that are T1D biomarker candidates. We performed an Ingenuity pathway analysis (Qiagen), which revealed an enrichment in proteins from cytokine/chemokine signaling (Fig. 5b). These results showed that plasma EVs carries several proteins that are candidates for T1D biomarkers, with a good likelihood to be developed in targeted proteomics assays and possible contributions to the disease process.
Evaluation of extracellular vesicle proteins potential as biomarker candidates. (a) Extracellular vesicle (EV) proteins from clusters 10 and 11 were compared the Human Plasma PeptideAtlas database, and a systematic review of type 1 diabetes (T1D) biomarker candidates30. (b) Network of T1D biomarker candidates present in EVs. The network was built after pathway-enrichment analysis performed with Ingenuity software (Qiagen).
Validation and biological activity of an EV protein identified in our meta-analysis
As the chemokine-signaling pathway was highly enriched in clusters 10 and 11 and among the plasma EV proteins that are T1D biomarker candidates (Figs. 4, 5b), we decided to further investigate this pathway. We were especially interested in this pathway as many chemokines play a role in T1D development31. A detailed annotation of this pathway revealed a total of 7 chemokines and 5 receptors detected, along with complete signaling branches related to the JAK/STAT and SRC/DOCK2/RAC1-2 cascades (Fig. 6a). Among the identified EV-chemokines, proplatelet basic protein (PPBP, also known as chemokine CXCL7) (Figs. 5b, 6a) has been validated as a T1D biomarker, with reports of its elevated levels in the plasma prior to and at the onset of the disease32,33. One of PPBP’s receptors, CXCR2, was also present in the plasma EVs (Fig. 6a). Therefore, we hypothesize that PPBP might have a role in T1D development. We first validated the presence of PPBP in EVs using the ExoView technology. In this technology, EVs are captured in microchips with antibodies against CD9, CD63, and CD81 then specific EV proteins are imaged by immunostaining using a fluorescence microscope. As a positive control, we stained for CD81 and our target PPBP. The images show an example of EVs captured with anti-CD9 antibodies and stained for CD81 and PPBP. The results show that the majority of the captured EVs contained CD81, while only a small fraction contained PPBP (Fig. 6b). The quantification showed that despite the number of particles positive for PPBP being low in plasma, it is significantly higher than the negative control for EV capture with pre-immune mouse antibody (MIgG), confirming the presence of this protein in plasma EVs (Fig. 6c). We next sought to determine if this increased level of plasma PPBP has a role in regulating cytokine-mediated apoptosis, a key process during T1D development. We pre-treated MIN6 β cells and Raw 264.7 macrophages with recombinant PPBP, prior to exposure to a cytokine cocktail (interferon γ, interleukin β, and tumor necrosis factor α). We found that PPBP induces MIN6 apoptosis, but in contrast, PPBP reduces Raw 264.7 apoptosis (Fig. 6d,e). Overall, these results show a remarkable enrichment of the chemokine-signaling pathway in EVs. In addition, we validated the presence of PPBP in EVs, which was identified in our meta-analysis. Furthermore, PPBP might play a role in T1D development by differentially regulating apoptosis in β cells and macrophages, two cell types that play key roles in T1D development.
Chemokine signaling components in extracellular vesicles and their activity in β cells and macrophages. (a) Chemokine signaling pathway components detected in plasma extracellular vesicles (clusters 10 and 11). (b) Showing the representative ExoView image of extracellular vesicles captured from human plasma in the chip and stained for CD81 and proplatelet basic protein (PPBP). (c) The bar graph represents number of total extracellular vesicles captured in the ExoView chip, and subpopulations containing CD81 and PPBP (n = 3). Significance by 1-way ANOVA *p ≤ 0.05, **p ≤ 0.01, & ****p ≤ 0.0001 (d,e) The bar graphs represent fold change in apoptosis in Raw 264.7 and MIN6 cells pretreated with different concentrations of PPBP for 24 h and then treated with a cytokine cocktail (CT, 100 ng/mL IFN-γ, 10 ng/mL TNF-α, and 5 ng/mL IL-1β) vs. control (noCT). N = 4 ± SD, Significance by 2-way ANOVA *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 & ****p ≤ 0.0001.
Discussion
Obtaining highly pure EV preparations is a complex task. For instance, plasma lipoproteins have similar sizes, and some particles have similar densities as EVs, making it difficult to attain pure EVs with techniques based on size and density6,7,8. Another challenge is the unavoidable presence of a high abundance of plasma proteins, such as albumin. To address this challenge, multiple purification steps with albumin depletion columns have been explored34. However, multi-step purifications lead to substantial sample losses, with almost complete EV loss after 3 purification steps6. Therefore, there is a need to better understand the characteristics of different isolation methods so that optimal approaches can be incorporated in each study. By applying a meta-analysis to datasets refined the composition of plasma EVs, leading two clusters with 1717 proteins enriched in EV proteins. We validated these clusters by performing a systematic review of human EV proteins that were analyzed by immunogold electron microscopy. Our analysis also quantified differences in contaminants across these methods and, herein, we provide insights into strengths and weaknesses of each method.
Centrifugation-based purification methods had similar contamination profiles. Diluting the plasma before the centrifugation seems to have a strong contaminant-reducing effect, through diminishing the concentration of proteins and consequently, their aggregation or absorption to EVs. However, the larger volume in DUC also affected the recovery of EVs during centrifugation. Inclusion of purification steps based on the same physical properties (e.g., ultracentrifugation vs. cushion ultracentrifugation) provided no added benefit in sample purity. Sequential purification steps had better results when using different physical-chemical properties, such as SEC followed by DGUC, but they led to a low EV recovery (~1%)11. Polymer-based precipitation (PP) had large amounts of lipoprotein contamination, probably due to their mechanism of enriching for EVs by binding to lipids. These findings suggest that understanding of contaminant characteristics is key to understanding limitations of each isolation technique, ways to improve sample preparation quality, and ability to distinguish activities of EVs from those of contaminants. In terms of EV sample recovery, high speed centrifugations had a low recovery, probably due to aggregation or disruption of EVs35,36. SEC had the best performance recovering proteins from clusters 10 and 11 combined, which is consistent with previous observation that SEC can recover 35% of the plasma EVs11. Centrifugation at 10,000xg showed an enrichment to cluster 11, probably by enriching for EVs with higher density. Therefore, for applications where pure materials are not required, such as targeted proteomics analysis, a single-step enrichment might be more appropriate than attempts to obtain highly purified materials in trace amounts.
Cell-specific proteins were identified in EVs from a variety of immune cells including lymphocytes, monocytes, neutrophils, and dendritic cells. These were accompanied by the presence of cell-surface CD antigens, which can be used as cellular markers. Many of these CD antigens have roles in cell signaling and cell-to-cell communication, consistent with earlier reports of EV participation in those events. A classic example of intercell communication by EVs is antigen presentation and T-cell activation by B-cell derived EVs37. Our pathway analysis further showed that plasma extracellular vesicles are also enriched in cell-to-cell communication and signaling proteins. However, the extent of the enrichment and the completeness of some signaling pathways were unprecedented. For instance, we found that the EV clusters had 133 CD antigens and complete signaling pathways. This opens the possibility that EVs can not only be a signaling messenger but complement cells that lack specific signaling pathways.
As a proof of principle for how these data can be applied, we also investigated the possibility of targeting EV proteins as T1D biomarkers and found that 41 current T1D biomarker candidates are present in EVs. Among these proteins is PPBP, which has been shown to be a T1D biomarker candidate in two separate studies32,33. We found that PPBP regulates apoptosis of β cells and macrophages. However, its role in T1D development is still poorly understood. The protection of macrophages and induction of β-cell apoptosis might represent a signal amplification mechanism, leading to an increase in β-cell death. The PPBP cleavage product, CXCL7, is a major chemokine that primes neutrophil migration by aggregating with platelets and inducing the formation of neutrophil extracellular traps (NETs)38. Neutrophil-platelet aggregates have been found to be abundant in blood of pre-diabetic and recent T1D onset mice and humans39. In addition, NETs were shown to cause β-cell death in vitro39, further supporting that PPBP might have a role in T1D development. However, additional work is needed to have a mechanistic understanding on the action of PPBP in T1D.
Overall, we provided an alternative approach for characterizing the EV composition. We found that C and SEC methods led to the best plasma EV recovery, but none of the studied methods led to pure preparations. Our analysis showed that plasma EVs are derived from a variety of cells and that they are enriched in cell surface markers and cell-to-cell communication molecules. Our study provides lists of proteins that are highly likely to be bona fide EV proteins than can be used for prioritization for mechanistic and biomarker studies. As an example, we validated the presence of PPBP in EVs and further showed that controls apoptosis in culture β cells and macrophages and might have a role in T1D development.
Data availability
Additional data are available in Open Science Framework28. This includes MaxQuant results and parameter files for each dataset under the folder “MaxQuant_results_and_parameters”, which are named based on their Pride accession numbers; an excel file containing the complete abundance data matrix under the folder “Processed_data_matrix”; and the results from the DAVID functional enrichment analysis under the folder “Enrichment analyses”.
Code availability
The R and Python scripts written to generate Fig. 5 and Figure S2 are available in GitHub23.
References
Raposo, G. & Stoorvogel, W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200, 373–383, https://doi.org/10.1083/jcb.201211138 (2013).
Buzas, E. I. The roles of extracellular vesicles in the immune system. Nat Rev Immunol, 1–15 https://doi.org/10.1038/s41577-022-00763-8 (2022).
Aguirre, R. S. et al. Extracellular vesicles in beta cell biology: Role of lipids in vesicle biogenesis, cargo, and intercellular signaling. Mol Metab 63, 101545, https://doi.org/10.1016/j.molmet.2022.101545 (2022).
Salomon, C. et al. Extracellular Vesicles and Their Emerging Roles as Cellular Messengers in Endocrinology: An Endocrine Society Scientific Statement. Endocr Rev 43, 441–468, https://doi.org/10.1210/endrev/bnac009 (2022).
Baranyai, T. et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS One 10, e0145686, https://doi.org/10.1371/journal.pone.0145686 (2015).
Brennan, K. et al. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci Rep 10, 1039, https://doi.org/10.1038/s41598-020-57497-7 (2020).
Burton, J. B., Carruthers, N. J. & Stemmer, P. M. Enriching extracellular vesicles for mass spectrometry. Mass Spectrom Rev https://doi.org/10.1002/mas.21738 (2021).
Coumans, F. A. W. et al. Methodological Guidelines to Study Extracellular Vesicles. Circ Res 120, 1632–1648, https://doi.org/10.1161/CIRCRESAHA.117.309417 (2017).
Thery, C. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7, 1535750, https://doi.org/10.1080/20013078.2018.1535750 (2018).
Xu, R., Greening, D. W., Rai, A., Ji, H. & Simpson, R. J. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 87, 11–25, https://doi.org/10.1016/j.ymeth.2015.04.008 (2015).
Vergauwen, G. et al. Robust sequential biophysical fractionation of blood plasma to study variations in the biomolecular landscape of systemically circulating extracellular vesicles across clinical conditions. J Extracell Vesicles 10, e12122, https://doi.org/10.1002/jev2.12122 (2021).
Lakhter, A. J. et al. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia 61, 1124–1134, https://doi.org/10.1007/s00125-018-4559-5 (2018).
Javeed, N. et al. Pro-inflammatory beta cell small extracellular vesicles induce beta cell failure through activation of the CXCL10/CXCR3 axis in diabetes. Cell Rep 36, 109613, https://doi.org/10.1016/j.celrep.2021.109613 (2021).
Zhu, X. et al. Exosomal miR-140-3p and miR-143-3p from TGF-beta1-treated pancreatic stellate cells target BCL2 mRNA to increase beta-cell apoptosis. Mol Cell Endocrinol 551, 111653, https://doi.org/10.1016/j.mce.2022.111653 (2022).
Shorten, A. & Shorten, B. What is meta-analysis? Evid Based Nurs 16, 3–4, https://doi.org/10.1136/eb-2012-101118 (2013).
Page, M. J. et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372, n71, https://doi.org/10.1136/bmj.n71 (2021).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50, D543–D552, https://doi.org/10.1093/nar/gkab1038 (2022).
Choi, M. et al. MassIVE.quant: a community resource of quantitative mass spectrometry-based proteomics datasets. Nat Methods 17, 981–984, https://doi.org/10.1038/s41592-020-0955-0 (2020).
Schwanhausser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342, https://doi.org/10.1038/nature10098 (2011).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372, https://doi.org/10.1038/nbt.1511 (2008).
Chu, V. T., Gottardo, R., Raftery, A. E., Bumgarner, R. E. & Yeung, K. Y. MeV+R: using MeV as a graphical user interface for Bioconductor applications in microarray analysis. Genome Biol 9, R118, https://doi.org/10.1186/gb-2008-9-7-r118 (2008).
Pathan, M. et al. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res 47, D516–D519, https://doi.org/10.1093/nar/gky1029 (2019).
Vallejo, M. C. Plasma Extracellular Vesicles. GitHub https://doi.org/10.5281/zenodo.10079817 (2023).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57, https://doi.org/10.1038/nprot.2008.211 (2009).
Jiang, L. et al. A Quantitative Proteome Map of the Human Body. Cell 183, 269–283 e219, https://doi.org/10.1016/j.cell.2020.08.036 (2020).
Junker, B. H., Klukas, C. & Schreiber, F. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7, 109, https://doi.org/10.1186/1471-2105-7-109 (2006).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, https://doi.org/10.1101/gr.1239303 (2003).
Vallejo, M. C. Human Plasma Extracellular Vesicle Proteomics Meta-analysis. Open Science Framework https://doi.org/10.17605/OSF.IO/2UQPK (2022).
Deutsch, E. W. et al. Advances and Utility of the Human Plasma Proteome. J Proteome Res 20, 5241–5263, https://doi.org/10.1021/acs.jproteome.1c00657 (2021).
Sarkar, S. et al. Systematic review of type 1 diabetes biomarkers reveals regulation in circulating proteins related to complement, lipid metabolism, and immune response. Clin Proteomics 20, 38, https://doi.org/10.1186/s12014-023-09429-6 (2023).
Eizirik, D. L., Colli, M. L. & Ortis, F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat Rev Endocrinol 5, 219–226, https://doi.org/10.1038/nrendo.2009.21 (2009).
Zhang, Q. et al. Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. J Exp Med 210, 191–203, https://doi.org/10.1084/jem.20111843 (2013).
Frohnert, B. I. et al. Predictive Modeling of Type 1 Diabetes Stages Using Disparate Data Sources. Diabetes 69, 238–248, https://doi.org/10.2337/db18-1263 (2020).
Palviainen, M. et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo-Implications for biomarker discovery. PLoS One 15, e0236439, https://doi.org/10.1371/journal.pone.0236439 (2020).
Linares, R., Tan, S., Gounou, C., Arraud, N. & Brisson, A. R. High-speed centrifugation induces aggregation of extracellular vesicles. J Extracell Vesicles 4, 29509, https://doi.org/10.3402/jev.v4.29509 (2015).
Nordin, J. Z. et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine 11, 879–883, https://doi.org/10.1016/j.nano.2015.01.003 (2015).
Raposo, G. et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med 183, 1161–1172, https://doi.org/10.1084/jem.183.3.1161 (1996).
Page, C. & Pitchford, S. Neutrophil and platelet complexes and their relevance to neutrophil recruitment and activation. Int Immunopharmacol 17, 1176–1184, https://doi.org/10.1016/j.intimp.2013.06.004 (2013).
Popp, S. K. et al. Circulating platelet-neutrophil aggregates characterize the development of type 1 diabetes in humans and NOD mice. JCI Insight 7, https://doi.org/10.1172/jci.insight.153993 (2022).
Vanderboom, P. M. et al. A size-exclusion-based approach for purifying extracellular vesicles from human plasma. Cell Rep Methods 1, https://doi.org/10.1016/j.crmeth.2021.100055 (2021).
Vanderboom, P. & Nair, K. S. Purification of human plasma exosomes for proteomics: optimization and application to detect changes in response to exercise. PRIDE Archive. https://identifiers.org/pride.project:PXD026483 (2021).
Han, Z. et al. Matrix-assisted laser desorption ionization mass spectrometry profiling of plasma exosomes evaluates osteosarcoma metastasis. iScience 24, 102906, https://doi.org/10.1016/j.isci.2021.102906 (2021).
Han, Z. & Qiao, L. Plasma-derived exosomes from healthy and osteosarcoma. PRIDE Archive. https://identifiers.org/pride.project:PXD024072 (2021).
Tunset, M. E. et al. Extracellular vesicles in patients in the acute phase of psychosis and after clinical improvement: an explorative study. PeerJ 8, e9714, https://doi.org/10.7717/peerj.9714 (2020).
Haslene-Hox, H. Human plasma extracellular vesicles LC-MS/MS. PRIDE Archive. https://identifiers.org/pride.project:PXD016293 (2020).
Hoshino, A. et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 182, 1044–1061 e1018, https://doi.org/10.1016/j.cell.2020.07.009 (2020).
Molina, H. & Lyden, D. Tissue- and plasma-derived exosomal protein biomarkers define multiple human cancers. PRIDE Archive. https://identifiers.org/pride.project:PXD018301 (2020).
Zhang, X., Borg, E. G. F., Liaci, A. M., Vos, H. R. & Stoorvogel, W. A novel three step protocol to isolate extracellular vesicles from plasma or cell culture medium with both high yield and purity. J Extracell Vesicles 9, 1791450, https://doi.org/10.1080/20013078.2020.1791450 (2020).
Vos, H. R. & Stoorvogel, W. A novel and simple strategy to isolate extracellular vesicles from human plasma and tissue culture medium with high yield and purity. PRIDE Archive. https://identifiers.org/pride.project:PXD015283 (2021).
Haderk, F. et al. Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci Immunol 2 https://doi.org/10.1126/sciimmunol.aah5509 (2017).
Iskar, M. & Seiffert, M. Proteome profiling of blood plasma-derived exosomes in chronic lymphocytic leukemia. PRIDE Archive. https://identifiers.org/pride.project:PXD004420 (2017).
Gallart-Palau, X. et al. Extracellular vesicles are rapidly purified from human plasma by PRotein Organic Solvent PRecipitation (PROSPR). Sci Rep 5, 14664, https://doi.org/10.1038/srep14664 (2015).
Gallart-Palau, X. & Sze, S. K. Isolation of extracellular vesicles by PROSPR. PRIDE Archive. https://identifiers.org/pride.project:PXD002668 (2015).
Acknowledgements
The authors thank Mr. Nathan Johnson of PNNL for his help in drafting Figs. 1, 6A. This work was supported by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant U01 DK127786 (to R.G.M, T.O.M. and S.R.), R01 DK060581 (to R.G.M), R01 DK133881 (to E.K.S. and R.G.M.), R01 DK121929 (to E.K.S.) and R01 DK126444 (to S.R.). S.R. was also supported by UAB-DRC P&F, CDIB Department, and UAB-Comprehensive Diabetes Center.
Author information
Authors and Affiliations
Contributions
M.C.V., S.H.P., S.R., E.K.S., T.O.M., R.G.M. and E.S.N. designed research study; M.V.C. performed the meta-analysis; F.H., S.S., E.C.E. and H.R.H. performed the experiments and analyzed respective data; E.C.E., S.M.P., S.S., I.D.L., Y.Y. and E.S.N. performed the systematic review; S.H.P., S.R., E.K.S., T.O.M., R.G.M. and E.S.N. provided guidance and resources to the project. M.C.V. and E.S.N. wrote the manuscript. All authors revised the manuscript and gave final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vallejo, M.C., Sarkar, S., Elliott, E.C. et al. A proteomic meta-analysis refinement of plasma extracellular vesicles. Sci Data 10, 837 (2023). https://doi.org/10.1038/s41597-023-02748-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02748-1