Abstract
The range of applications of digital surface models of the bones in science and industry is wide. Three-dimensional reconstructions of bones are used in biomechanics, biomedical engineering, medical image processing, orthopedics, traumatology, radiology, patient education, anatomy, anthropometry, forensic anthropology, ergonomics, usability and human factors engineering, or accident and injury analysis and prevention. No open access database or repository of skeletal surface models of the full lower extremities exists. Therefore, the objective of this publication was to provide access to consistent complete bone models of the pelvis and lower limbs of multiple subjects, including biometric data. Segmentations and surface models of the bones of the lower extremities of more than twenty subjects were created from open access postmortem whole-body computed tomography scans. The database provides a broad range of applications by giving access to the data of the complete process chain, from the raw medical imaging data through the segmentations to the surface models.
Similar content being viewed by others
Background & Summary
The field of application of digital bone models is broad. Three-dimensional (3D) reconstructions of bones are used in biomechanics, biomedical engineering and medical image processing for musculoskeletal modelling1,2, finite element analyses3, statistical shape modelling4,5,6 or 3D reconstruction from sparse imaging data, such as radiographs7,8 or EOS images9. 3D reconstructions of the bones are used in orthopedics, traumatology or radiology for the development of implants10,11,12,13,14, surgical instruments15,16 or procedures, for diagnosis and decision-making17,18, preoperative planning19,20 and navigational guidance during computer assisted surgery8,21, the evaluation of outcome22, surgery simulation23, surgical education and training24, especially in the context of personalized, patient-specific, customized or individualized medicine. The surgical guidance based on bone models can be virtual, augmented25 or mixed reality26, or 3D printed27,28. Further fields of application are anatomy and patient education29,30, morphometrics31 and anthropometry32,33, forensic anthropology34,35, ergonomics, usability and human factors engineering36, accident and injury analysis and prevention37.
However, to the best of the author’s knowledge, no open access database or repository of skeletal surface models of the full lower extremities exists. Therefore, the objective of this study was to provide access to consistent complete bone models of the pelvis and lower limbs of multiple subjects. The database is supposed to enable other researches to quickly develop, test and verify new methods, approaches, algorithms or proofs of concept without the time-consuming and labor-intensive work of data collection and curation, segmentation and reconstruction. The database is expected to help the scientific community to facilitate research and improve the reproducibility and comparability of studies by giving access to the raw medical imaging data, including the metadata of the subjects and the segmentations and surface models of the bones. Hence, different researchers and research groups can resort to the same datasets for the validation of methods and comparison of results. Different deep learning models for artificial intelligence-based bone reconstruction, for instance, could be benchmarked by applying them to the raw computed tomography (CT) data and comparing the automatic with the manual segmentations of the database. The database can also be used as additional training data for existing deep learning models38,39.
Methods
Source of the raw CT data
The segmentations and models of the bones of the lower extremities were created from anonymized postmortem CT scans of the whole body originally published by Kistler et al. in the Swiss Institute for Computer Assisted Surgery Medical Image Repository (smir.ch) as open access Virtual Skeleton Database (VSD)40. The CT datasets were provided by the forensic institutes of the universities of Bern and Zürich and shared under the Creative Commons Attribution-NonCommercial-ShareAlike (CC BY-NC-SA) license after ethical approval of the Cantonal Ethics Committee Bern41. Further information about the datasets can be found in the literature cited40,41. Due to ongoing difficulties in accessing the SMIR website, the author decided to reupload the original datasets without any changes to the open access hosting service Zenodo: https://doi.org/10.5281/zenodo.827036442.
CAUTION
The VSD contains a few inconsistencies, such as duplicate CT datasets. The author of this publication is not connected to the SMIR or VSD and, therefore, not responsible for errors in the VSD. However, errors that the author recognized during the work with the VSD were logged and are reported in the reupload of the VSD42.
Subject selection
Twenty subjects (ten male and ten female) were selected from the VSD for the creation of the bone models with the objective of covering a wide age range.
The inclusion criteria were:
-
Availability of age, body weight and body height.
-
Integrity and completeness of the lower body’s skeletal anatomy.
The exclusion criteria were:
-
Difference between the gender specified in the metadata and the biological sex visible in the CT data.
-
Presence of artificial joints or bone fractures.
The average age, weight and height of the twenty subjects were 52 ± 21 years, 70 ± 13 kg and 1.7 ± 0.1 m, respectively. An overview of the subjects is presented in Table 1. Some subjects were processed before the inclusion and exclusion criteria were defined. Ten of the subjects did not meet the criteria. These ten additional subjects are also published as part of the database since they still might be useful for some applications, but they are tagged by a comment in the database so they can be easily identified by the user (see Table 1).
Reconstruction of the osseous anatomy
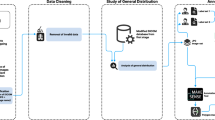
The bone surfaces were semi-automatically reconstructed by thresholding (Fig. 1). Two hundred Hounsfield units43 were chosen as the lower threshold and the maximum Hounsfield unit value present in the volume data was selected as the upper threshold. Subsequently, a manual post-processing using the software 3D Slicer (slicer.org) with default smoothing settings was performed44. The bones were manually segmented at the joints if necessary. All joints were segmented. However, some segments contain multiple components as follows:
-
Sacrum including the coccyx (if not fused with the sacrum)
-
Hip bone (also called pelvic, innominate or coxal bone)
-
Femur
-
Patella
-
Tibia
-
Fibula
-
Talus
-
Calcaneus
-
Tarsals, including the cuboid, navicular and three cuneiforms
-
Metatarsals
-
Phalanges
Separate segments were created for the left and right leg. Some segments contain small sesamoid bones if present. This applies to the metatarsals for all subjects but, in some cases, also to other bones, such as the femurs.
After the segmentation, the bones were reconstructed by manually closing holes present in the outer surface. No gap closing, hole filling or wrapping algorithms were used. The reconstructed surface models were exported as mesh files in the Polygon File Format (PLY) and imported into MATLAB using a conservative decimation and remeshing procedure (Fig. 1). The Hausdorff distance between input and output mesh was limited to 0.05 mm for the decimator. The adaptive remesher permitted a maximum deviation of 0.05 mm from the input mesh with a minimum and maximum edge length of 0.5 and 100 mm, respectively. The decimator and remesher are plugins of the software OpenFlipper (openflipper.org)45.
CAUTION
Each reconstruction of anatomical structures from medical images is subject to cumulative spatial errors arising from each step of the process chain. While the section “Technical Validation” should give an impression of the error that can be expected from the workflow described, users of the database should take into account the risk of larger reconstruction errors depending on the application intended.
The bone models of each subject can be visualized by running the MATLAB or Python examples. One subject is presented in Fig. 2. The 3D reconstructions were created by the author as a private side project between 2017 and 2022. Parts of the database containing fewer subjects and only the pelvis and femurs were published previously as part of other studies of the author46,47. This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Analysis of the surface models stored as MAT files
The database was searched for duplicate subjects using a two-stage registration process. Each pelvis was transformed into an automatically detected pelvic coordinate system based on the anterior pelvic plane using the iterative tangential plane method46. Subsequently, the sacrum of each subject was registered to the sacra of all other subjects using a rigid iterative closest points algorithm. Lower outliers of the root mean square error between the two registered sacra were examined. One duplicate subject was identified, excluded from the database and replaced by another subject.
Each bone model of all subjects was visually reviewed for internal cavities connected to the outer surface or connections between the inner and outer surface, and corrections were performed if necessary. The mesh topology was checked for the following errors using MATLAB:
-
Duplicate, non-manifold and unreferenced vertices.
-
Boundary, non-manifold and conflictingly oriented edges.
-
Duplicate and degenerated faces.
-
Self-intersections and intersections with adjacent bones.
The errors were corrected if present.
The volume enclosed by the outer surface of the bone models was calculated and is presented in Table 2. The values were compared with those from literature. However, caution must be applied since different definitions and measurement methods of the bone volume exist. Studies reporting the trabecular or cortical volume of the bones were not considered. The values of the bone volume correspond to those observed in previous studies48,49,50,51.
Data Records
As mentioned above, a mirror of the complete VSD as hosted originally by Kistler et al. at smir.ch is available at Zenodo: https://doi.org/10.5281/zenodo.827036442.
The CT volume data, segmentations, reconstructions and raw PLY mesh files of the subjects of Table 1 are accessible via Zenodo: https://doi.org/10.5281/zenodo.830244852. The files of each subject are linked by a project file, called MRML scene file, that can be opened with the open-source medical imaging software 3D Slicer (slicer.org).
The post-processed mesh files of the subjects of Table 1 are stored as MATLAB MAT files, released as Git repository at https://github.com/MCM-Fischer/VSDFullBodyBoneModels and versioned via Zenodo: https://doi.org/10.5281/zenodo.831673053. The use of the MAT files is explained by examples for MATLAB and Python in the Git repository.
Technical Validation
The VSD also contains CT data of the European Spine Phantom that was introduced by Kalender et al. in 199554. The CT phantom data was used to evaluate the reconstruction process described above. After the creation of the surface model of the phantom, landmarks and areas were manually selected on the surface model of the phantom. Planes or cylinders were fitted to the areas selected to calculate the geometric parameters of the phantom. The errors between the reconstructed and the reference values of the geometric parameters reported in the publication by Kalender et al. are presented in Table 3. The mean error was 0.2 ± 0.4 mm and the mean absolute error was 0.4 ± 0.2 mm. This agrees well with accuracies reported in literature for 3D bone reconstruction using CT. Lalone et al. reported a mean error of 0.4 ± 0.3 mm for the cortical bone of the upper extremities55, Wang et al. reported a mean error of 0.5 ± 0.2 mm for machined bone specimens from the femur and tibia56 and van den Broeck et al. reported a mean absolute error of 0.5 ± 0.2 mm for the tibia57.
Code availability
The code used to create and analyze the datasets is openly accessible via https://github.com/MCM-Fischer/VSDFullBodyBoneModels and versioned at Zenondo: https://doi.org/10.5281/zenodo.831673053.
References
Suwarganda, E. K. et al. Minimal medical imaging can accurately reconstruct geometric bone models for musculoskeletal models. PLoS One 14, e0205628, https://doi.org/10.1371/journal.pone.0205628 (2019).
Modenese, L. & Renault, J.-B. Automatic generation of personalised skeletal models of the lower limb from three-dimensional bone geometries. J. Biomech. 116, 110186, https://doi.org/10.1016/j.jbiomech.2020.110186 (2021).
Burastero, G. et al. Use of porous custom-made cones for meta-diaphyseal bone defects reconstruction in knee revision surgery: a clinical and biomechanical analysis. Arch. Orthop. Trauma Surg. 140, 2041–2055, https://doi.org/10.1007/s00402-020-03670-6 (2020).
Grant, T. M. et al. Development and validation of statistical shape models of the primary functional bone segments of the foot. PeerJ 8, e8397, https://doi.org/10.7717/peerj.8397 (2020).
Nolte, D., Ko, S.-T., Bull, A. M. & Kedgley, A. E. Reconstruction of the lower limb bones from digitised anatomical landmarks using statistical shape modelling. Gait Posture 77, 269–275, https://doi.org/10.1016/j.gaitpost.2020.02.010 (2020).
Ahrend, M.-D. et al. Development of generic Asian pelvic bone models using CT-based 3D statistical modelling. J. Orthop. Translat. 20, 100–106, https://doi.org/10.1016/j.jot.2019.10.004 (2020).
Ün, M. K., Avşar, E. & Akçalı, İ. D. An analytical method to create patient-specific deformed bone models using X-ray images and a healthy bone model. Comput. Biol. Med. 104, 43–51, https://doi.org/10.1016/j.compbiomed.2018.11.003 (2019).
Massé, V. & Ghate, R. S. Using standard X-ray images to create 3D digital bone models and patient-matched guides for aiding implant positioning and sizing in total knee arthroplasty. Comput. Assist. Surg. 26, 31–40, https://doi.org/10.1080/24699322.2021.1894239 (2021).
Yan, W. et al. Femoral and tibial torsion measurements based on EOS imaging compared to 3D CT reconstruction measurements. Ann. Transl. Med. 7, 460, https://doi.org/10.21037/atm.2019.08.49 (2019).
Belvedere, C. et al. New comprehensive procedure for custom-made total ankle replacements: Medical imaging, joint modeling, prosthesis design, and 3D printing. J. Orthop. Res. 37, 760–768, https://doi.org/10.1002/jor.24198 (2019).
Schmutz, B., Rathnayaka, K. & Albrecht, T. Anatomical fitting of a plate shape directly derived from a 3D statistical bone model of the tibia. J. Clin. Orthop. Trauma 10, S236–S241, https://doi.org/10.1016/j.jcot.2019.04.019 (2019).
Dupraz, I. et al. Using statistical shape models to optimize TKA implant design. Appl. Sci. 12, 1020, https://doi.org/10.3390/app12031020 (2022).
Thiesen, D. M. et al. Femoral antecurvation - a 3D CT analysis of 1232 adult femurs. PLoS One 13, e0204961, https://doi.org/10.1371/journal.pone.0204961 (2018).
Grothues, S. A. G. A. & Radermacher, K. Variation of the three-dimensional femoral J-curve in the native knee. J. Pers. Med. 11, 592, https://doi.org/10.3390/jpm11070592 (2021).
Mattei, L., Pellegrino, P., Calò, M., Bistolfi, A. & Castoldi, F. Patient specific instrumentation in total knee arthroplasty: a state of the art. Ann. Transl. Med. 4, 126, https://doi.org/10.21037/atm.2016.03.33 (2016).
Cartiaux, O., Paul, L., Francq, B. G., Banse, X. & Docquier, P.-L. Improved accuracy with 3D planning and patient-specific instruments during simulated pelvic bone tumor surgery. Ann. Biomed. Eng. 42, 205–213, https://doi.org/10.1007/s10439-013-0890-7 (2014).
Morton, A. M., Akhbari, B., Moore, D. C. & Crisco, J. J. Osteophyte volume calculation using dissimilarity-excluding Procrustes registration of archived bone models from healthy volunteers. J. Orthop. Res. 38, 1307–1315, https://doi.org/10.1002/jor.24569 (2020).
DeFroda, S. F. et al. Quantification of acetabular coverage on 3-dimensional reconstructed computed tomography scan bone models in patients with femoroacetabular impingement syndrome: a descriptive study. Orthop. J. Sports Med. 9, 23259671211049457, https://doi.org/10.1177/23259671211049457 (2021).
Habor, J., Fischer, M. C. M., Tokunaga, K., Okamoto, M. & Radermacher, K. The patient-specific combined target zone for morpho-functional planning of total hip arthroplasty. J. Pers. Med. 11, 817, https://doi.org/10.3390/jpm11080817 (2021).
Palit, A., King, R., Pierrepont, J. & Williams, M. A. Development of bony range of motion (B-ROM) boundary for total hip replacement planning. Comput. Methods Programs Biomed. 222, 106937, https://doi.org/10.1016/j.cmpb.2022.106937 (2022).
Nakano, N., Audenaert, E., Ranawat, A. & Khanduja, V. Review: current concepts in computer-assisted hip arthroscopy. Int. J. Med. Robot. 14, e1929, https://doi.org/10.1002/rcs.1929 (2018).
Guidetti, M. et al. MRI- and CT-based metrics for the quantification of arthroscopic bone resections in femoroacetabular impingement syndrome. J. Orthop. Res. 40, 1174–1181, https://doi.org/10.1002/jor.25139 (2022).
Stefan, P. et al. Three-dimensional printed computed tomography-based bone models for spine surgery simulation. Simul. Healthc. 15, 61–66, https://doi.org/10.1097/SIH.0000000000000417 (2020).
Radetzki, F. et al. Potentialities and limitations of a database constructing three-dimensional virtual bone models. Surg. Radiol. Anat. 35, 963–968, https://doi.org/10.1007/s00276-013-1118-0 (2013).
Schlueter-Brust, K. et al. Augmented-reality-assisted K-wire placement for glenoid component positioning in reversed shoulder arthroplasty: a proof-of-concept study. J. Pers. Med. 11, https://doi.org/10.3390/jpm11080777 (2021).
Wei, P. et al. Percutaneous kyphoplasty assisted with/without mixed reality technology in treatment of OVCF with IVC: a prospective study. J. Orthop. Surg. Res. 14, 255, https://doi.org/10.1186/s13018-019-1303-x (2019).
Brinkmann, E. J. & Fitz, W. Custom total knee: understanding the indication and process. Arch. Orthop. Trauma Surg. 141, 2205–2216, https://doi.org/10.1007/s00402-021-04172-9 (2021).
Spencer-Gardner, L. et al. Patient-specific instrumentation improves the accuracy of acetabular component placement in total hip arthroplasty. Bone Joint J. 98-B, 1342–1346, https://doi.org/10.1302/0301-620X.98B10.37808 (2016).
Lozano, M. T. U. et al. A study evaluating the level of satisfaction of the students of health sciences about the use of 3D printed bone models. In Proceedings of the Sixth International Conference on Technological Ecosystems for Enhancing Multiculturality (ed. García-Peñalvo, F. J.), 368–372, https://doi.org/10.1145/3284179.3284242 (ACM, 2018).
Stephens, M. H. et al. 3-D bone models to improve treatment initiation among patients with osteoporosis: A randomised controlled pilot trial. Psychol. Health 31, 487–497, https://doi.org/10.1080/08870446.2015.1112389 (2016).
Nozaki, S., Watanabe, K., Kamiya, T., Katayose, M. & Ogihara, N. Three‐dimensional morphological variations of the human calcaneus investigated using geometric morphometrics. Clin. Anat. 33, 751–758, https://doi.org/10.1002/ca.23501 (2020).
Chui, C. S. et al. Population-based and personalized design of total knee replacement prosthesis for additive manufacturing based on Chinese anthropometric data. Engineering 7, 386–394, https://doi.org/10.1016/j.eng.2020.02.017 (2021).
Bachmeier, A. T., Euler, E., Bader, R., Böcker, W. & Thaller, P. H. Novel method for determining bone dimensions relevant for longitudinal and transverse distraction osteogenesis and application in the human tibia and fibula. Ann. Anat. 234, 151656, https://doi.org/10.1016/j.aanat.2020.151656 (2021).
Colman, K. L. et al. The accuracy of 3D virtual bone models of the pelvis for morphological sex estimation. Int. J. Legal Med. 133, 1853–1860, https://doi.org/10.1007/s00414-019-02002-7 (2019).
Colman, K. L. et al. Virtual forensic anthropology: the accuracy of osteometric analysis of 3D bone models derived from clinical computed tomography (CT) scans. Forensic Sci. Int. 304, 109963, https://doi.org/10.1016/j.forsciint.2019.109963 (2019).
Kermavnar, T., Shannon, A. & O’Sullivan, L. W. The application of additive manufacturing/3D printing in ergonomic aspects of product design: A systematic review. Appl. Ergon. 97, 103528, https://doi.org/10.1016/j.apergo.2021.103528 (2021).
Li, G., Yang, J. & Simms, C. The influence of gait stance on pedestrian lower limb injury risk. Accid. Anal. Prev. 85, 83–92, https://doi.org/10.1016/j.aap.2015.07.012 (2015).
Liu, P. et al. Deep learning to segment pelvic bones: large-scale CT datasets and baseline models. Int. J. Comput. Assist. Radiol. Surg. 16, 749–756, https://doi.org/10.1007/s11548-021-02363-8 (2021).
Li, Z. et al. Deep learning approach for guiding three-dimensional computed tomography reconstruction of lower limbs for robotically-assisted total knee arthroplasty. Int. J. Med. Robot. 17, e2300, https://doi.org/10.1002/rcs.2300 (2021).
Kistler, M., Bonaretti, S., Pfahrer, M., Niklaus, R. & Büchler, P. The virtual skeleton database: an open access repository for biomedical research and collaboration. J. Med. Internet Res. 15, e245, https://doi.org/10.2196/jmir.2930 (2013).
Kistler, M. A database framework to incorporate statistical variability in biomechanical simulations. PhD thesis. Faculty of Medicine of the University of Bern, https://boristheses.unibe.ch/916 (2014).
Kistler, M. VSDFullBody: The Virtual Skeleton Database Full Body CT Collection, Zenodo, https://doi.org/10.5281/zenodo.8270364 (2013).
Sugano, N. et al. Effects of CT threshold value to make a surface bone model on accuracy of shape-based registration in a CT-based navigation system for hip surgery. Int. Congr. Ser. 1230, 319–324, https://doi.org/10.1016/S0531-5131(01)00070-X (2001).
Fedorov, A. et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 30, 1323–1341, https://doi.org/10.1016/j.mri.2012.05.001 (2012).
Möbius, J. & Kobbelt, L. OpenFlipper: an open source geometry processing and rendering framework. In Curves and Surfaces. 7th International Conference, Avignon, France, June 2010, Revised Selected Papers (ed. Boissonnat, J.-D.), 488–500, https://doi.org/10.1007/978-3-642-27413-8_31 (Springer, 2012).
Fischer, M. C. M., Krooß, F., Habor, J. & Radermacher, K. A robust method for automatic identification of landmarks on surface models of the pelvis. Sci. Rep. 9, 13322, https://doi.org/10.1038/s41598-019-49573-4 (2019).
Fischer, M. C. M., Grothues, S. A. G. A., Habor, J., La Fuente, Mde & Radermacher, K. A robust method for automatic identification of femoral landmarks, axes, planes and bone coordinate systems using surface models. Sci. Rep. 10, 20859, https://doi.org/10.1038/s41598-020-77479-z (2020).
Shao, H., Chen, C., Scholl, D., Faizan, A. & Chen, A. F. Tibial shaft anatomy differs between Caucasians and East Asian individuals. Knee Surg. Sports Traumatol. Arthrosc. 26, 2758–2765, https://doi.org/10.1007/s00167-017-4724-2 (2018).
Hishmat, A. M. et al. Efficacy of automated three-dimensional image reconstruction of the femur from postmortem computed tomography data in morphometry for victim identification. Leg. Med. (Tokyo) 16, 114–117, https://doi.org/10.1016/j.legalmed.2014.01.004 (2014).
Zhan, M.-J. et al. Estimation of sex based on patella measurements in a contemporary Chinese population using multidetector computed tomography: An automatic measurement method. Leg. Med. (Tokyo) 47, 101778, https://doi.org/10.1016/j.legalmed.2020.101778 (2020).
Kuo, C.-C. et al. Three-dimensional computer graphics-based ankle morphometry with computerized tomography for total ankle replacement design and positioning. Clin. Anat. 27, 659–668, https://doi.org/10.1002/ca.22296 (2014).
Fischer, M. C. M. VSDFullBodyBoneReconstruction: Segmentations and surface models of bones of the entire lower body created from cadaver CT scans from the VSDFullBody collection. Version 1.0.0, Zenodo, https://doi.org/10.5281/zenodo.8302448 (2023).
Fischer, M. C. M. VSDFullBodyBoneModels: 3D surface models of the bones of the lower body created from CT datasets of the open access VSDFullBody collection. Version 3.0, Zenodo, https://doi.org/10.5281/zenodo.8302448 (2023).
Kalender, W. A. et al. The European Spine Phantom - a tool for standardization and quality control in spinal bone mineral measurements by DXA and QCT. Eur. J. Radiol. 20, 83–92, https://doi.org/10.1016/0720-048x(95)00631-y (1995).
Lalone, E. A., Willing, R. T., Shannon, H. L., King, G. J. W. & Johnson, J. A. Accuracy assessment of 3D bone reconstructions using CT: an intro comparison. Med. Eng. Phys. 37, 729–738, https://doi.org/10.1016/j.medengphy.2015.04.010 (2015).
Wang, J., Ye, M., Liu, Z. & Wang, C. Precision of cortical bone reconstruction based on 3D CT scans. Comput. Med. Imaging Graph. 33, 235–241, https://doi.org/10.1016/j.compmedimag.2009.01.001 (2009).
van den Broeck, J., Vereecke, E., Wirix-Speetjens, R. & Vander Sloten, J. Segmentation accuracy of long bones. Med. Eng. Phys. 36, 949–953, https://doi.org/10.1016/j.medengphy.2014.03.016 (2014).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
M.C.M.F.: Conceptualization, methodology, software, data curation, visualization and writing.
Corresponding author
Ethics declarations
Competing interests
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fischer, M.C.M. Database of segmentations and surface models of bones of the entire lower body created from cadaver CT scans. Sci Data 10, 763 (2023). https://doi.org/10.1038/s41597-023-02669-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02669-z