Abstract
Grand Canonical Monte Carlo is an important method for performing molecular-level simulations and assisting the study and development of nanoporous materials for gas capture applications. These simulations are based on the use of force fields and partial charges to model the interaction between the adsorbent molecules and the solid framework. The choice of the force field parameters and partial charges can significantly impact the results obtained, however, there are very few databases available to support a comprehensive impact evaluation. Here, we present a database of simulations of CO2 and N2 adsorption isotherms on 690 metal-organic frameworks taken from the CoRE MOF 2014 database. We performed simulations with two force fields (UFF and DREIDING), six partial charge schemes (no charges, Qeq, EQeq, MPNN, PACMOF, and DDEC), and three temperatures (273, 298, 323 K). The resulting isotherms compose the Charge-dependent, Reproducible, Accessible, Forcefield-dependent, and Temperature-dependent Exploratory Database (CRAFTED) of adsorption isotherms.
Similar content being viewed by others
Background & Summary
Carbon capture, storage, and utilization is considered as one of the key strategies required to reduce anthropogenic carbon dioxide emissions and their impacts on climate change1. The most viable option for this approach is to focus on capturing CO2 from point sources, such as fossil fuel power plants, fuel processing plants and other industrial plants where carbon capture technology can be applied to streams with industrial scale flow rates2. However, despite several decades of intensive research, carbon capture in an economically viable way remains an enormous challenge3.
Adsorption processes are considered to be a promising alternative to the conventional absorption processes due the their low regeneration energy, high selectivity, and high capture capacity4. Combined, these characteristics may lead to energy-efficient processes for industrial scale capture and utilization of greenhouse gases (GHG). At the heart of a typical adsorption process for gas separation, such as the Pressure Swing Adsorption process, is the active adsorbent material; and the efficiency of the process crucially depends on the properties of this material. Within the different adsorbent materials that are potentially available for this process, crystalline nanoporous materials such as metal-organic frameworks (MOF)5,6,7,8, covalent organic frameworks (COF)9,10,11, zeolitic imidazolate frameworks (ZIF)12,13,14, and zeolites15 feature many of the necessary characteristics for a solid sorbent for efficient gas separation under the conditions of interest.
These families of materials contain hundreds of thousands synthesized structures and countless more hypothetical ones, featuring pores of different size, shape, and chemical characteristics. This creates large exploration space for studies that seek to identify the best candidates for a given gas capture application. This endeavour, however, is not possible via a brute-force experimental campaign. The number of large databases built upon experimental16,17,18,19,20,21,22 and hypothetical23,24 structures, combined with the continuous growth of diversity and scope of new materials due to the advancements in digital reticular chemistry25,26, make high-throughput computational screening (HTCS) approaches an imperative strategy for efficient exploration of the vast chemical landscape of crystalline nanoporous adsorbents27,28.
Most of the HTCS studies for carbon capture and related problems are based on using Grand Canonical Monte Carlo (GCMC) simulations to generate adsorption data. This data is then used to form some simple material performance metrics or is passed on to the process level modelling to explore the performance of candidate materials under realistic process conditions.
To perform molecular simulations such as GCMC, one needs a set of parameters that describe the interactions among the adsorbate molecules, and between the adsorbate molecules and the atoms of the adsorbent material; this set of parameters is called a force field.
Over the years, many force fields have been developed for various purposes and several options are available to describe adsorption of gases such as carbon dioxide in materials such as MOFs. Invariably, the predicted equilibrium adsorption data and, consequently, the ranking of materials and the recommendations of the screening study will depend on the choice of the force field.
This poses several fundamental questions. How sensitive is the adsorption data to the choice of the force field parameters? How does this sensitivity vary across different categories or classes of materials? And ultimately, is a ranking of porous materials for a particular application a robust result or is it contingent on using a particular force field?
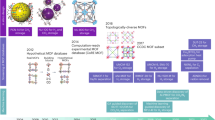
To start to explore these questions one needs a representative mass of adsorption data covering typical choices of the force field parameters, materials, gases and conditions. This defines the remit of the current article where we tasked ourselves with building such a database (or at least, the first block of conditions).
To explain the contents of the database and our approach, let us delve first into components of the classical force fields and the typical options available for the studies of adsorption of gases in MOFs and related materials. In the classical force fields, the non-bonded interactions are modeled as a sum of van der Waals and Coulomb potentials29. The van der Waals interactions between the adsorbed molecules and the framework are usually modeled by the Lennard-Jones (LJ) potential, which is an effective potential with two fitted parameters that can capture most of the intermolecular effects relevant to physisorption. The parameters for the atoms can be taken from generic force fields such as the Universal Force Field (UFF)30, DREIDING31 and TraPPE32, with the interactions between different atom species computed using mixing rules such as Lorentz-Berthelot33 or Jorgensen34.
The Coulombic interactions are modeled by partial atomic charges assigned to the atoms which need to be calculated for each material. There are several charge assignment methods available, and they can be divided into three main groups: i. methods derived from quantum chemistry calculations (e.g. RESP35, CHELP36, REPEAT37, and DDEC38,39,40); ii. methods based on charge equilibration (e.g. Qeq41, PQeq42, EQeq43, and FC-Qeq44); and iii. methods based on machine learning algorithms, such as PACMOF45 and MPNN46, that are trained to reproduce the quantum chemistry-based charges but at a much lower computational cost.
Lately, there have been several studies evaluating the accuracy of different methods for calculating partial atomic charges47,48,49,50,51,52, however, little is known about the combined impact of force field and partial charge selection on material-level analysis and its implication on process-level performance metrics. Despite the fact that the deficiencies of charge equilibration methods have already been recognised in the scientific literature, these methods are still commonly used in HTCS studies either as a pre-screening approach53,54 or as the final choice of charge assignment method in MOFs55,56,57 due to their simplicity and computational efficiency. Furthermore, the original Lennard-Jones force field parameters were derived and validated using specific partial charge schemes (Qeq30 for UFF and Gasteiger58 for DREIDING), thus the combination of these parameters with different charge assignment methodologies, even if more accurate, may not necessarily generate better results. Hence, none of the typical combinations of molecular-level modelling choices used in the simulation community (e.g., DREIDING + DDEC, UFF + DDEC, etc.) can be considered as systematically developed or validated and cannot be expected to produce accurate adsorption predictions. These considerations guide us on the choices of the parameters to consider in the database.
The database contains simulated adsorption isotherms for 690 MOFs selected from the CoRE MOF 201416,17 database. The simulations were performed for the adsorption of CO2 and N2 with two force fields (UFF and DREIDING), six partial charge schemes (no charge, Qeq, EQeq, PACMOF, MPNN, and DDEC), at three temperatures (273, 298, 323 K). The resulting isotherms compose the Charge-dependent, Reproducible, Accessible, Forcefield-dependent, and Temperature-dependent Exploratory Database (CRAFTED)59 of adsorption isotherms. CRAFTED provides a convenient platform to explore the sensitivity of simulation outcomes to molecular modeling choices at the material (structure-property relationship) and process levels (structure-property-performance relationship).
Methods
Structure selection
Starting from the 2932 structures present in the CoRE MOF 201416,17, first a set 726 structures that can be simultaneously modelled by both UFF and DREIDING force fields were selected. From these structures, 36 structures were removed due to the presence of unbound water molecules, counter-ions and/or hydrogen atom with incorrect bond lengths/angles. Therefore, a subset of 690 materials that can be modelled by all force fields and partial charge models was obtained and will be referred to as “CRAFTED structures”.
Partial charges calculation
The DDEC partial charges38,39,40 were taken without modification from the CoRE MOF 201416,17 database. The EQeq partial charges43 were calculated using the the extended charge equilibration method as implemented in the EQeq software v1.1.0 (https://github.com/danieleongari/EQeq). The Qeq partial charges41 were calculated using the default implementation available in RASPA60 v2.0.45. The PACMOF partial charges45 were calculated using the default Python implementation available in the PACMOF package (https://github.com/arung-northwestern/pacmof). The MPNN partial charges46 were calculated using the MPNN package (https://github.com/SimonEnsemble/mpn_charges).
Grand canonical Monte Carlo simulations
Atomistic Grand Canonical Monte Carlo (GCMC) simulations were performed using a force field-based algorithm as implemented in RASPA60,61 v2.0.45. Interaction energies between non-bonded atoms were computed through a combination of Lennard-Jones (LJ) and Coulomb potentials
where i and j are interacting atom indexes and rij is their interatomic distance. εij and σij are the well depth and diameter, respectively. The LJ parameters between atoms of different types were calculated using the Lorentz-Berthelot mixing rules
LJ parameters for framework atoms were taken from Universal Force Field (UFF)30 or DREIDING31 (see Table 1). The parameters for the adsorbed molecules were taken from the TraPPE32 force field (see Table 2). All simulations were performed with 10,000 Monte Carlo cycles. Swap (insertion or deletion with with a probability of 50% for each), translations, rotations, and re-insertions moves were tried with probabilities 0.5, 0.3, 0.1, and 0.1, respectively. To avoid the use of long initialization cycles, each isotherm was calculated in a single simulation, with each pressure point of the simulation starting from the result of the previous one. The uptake values for each pressure were obtained by averaging over the GCMC equilibrium phase, determined using the Marginal Standard Error Rule. For more information, please refer to section Automatic transient regime detection and truncation.
All atoms in the MOF were held fixed at their crystallographic positions. The number of unit cells used was different for each MOF to ensure that the perpendicular lengths of the supercell were greater than twice the cutoff used. The cutoff for Lennard-Jones and charge-charge short-range interactions was 12.8 Å and the Ewald sum technique was applied to compute the long-range electrostatic interactions with a relative precision of 10−6. The Lennard-Jones potential was shifted to zero at the cutoff. Fugacities needed to impose equilibrium between the system and the external ideal gas reservoir at each pressure were calculated using the Peng-Robinson equation of state62 with the critical parameters for each gas taken from Table 3. All GCMC uptake data report the absolute adsorption value in mol/kg units.
Lennard-Jones parameters
The Lennard-Jones parameters for DREIDING and UFF force fields used in the calculations for the framework atoms are shown in Table 1. For simplicity, only the atoms that are present in both UFF and DREIDING are shown. The TraPPE parameters used for the gas molecules are present in Table 2. The critical parameters used in the Peng-Robinson equation to calculate the fugacity are present in Table 3.
Automatic equilibration detection and truncation
To eliminate the use of long initialization cycles, the Marginal Standard Error Rule (MSER)63 was applied to automatically detect the ideal truncation point using the pyMSER package v1.0.18 (https://github.com/IBM/pymser), so that the averages were taken only over the equilibrated phase of the simulation. The output of this method is the equilibrated average of the observable, alongside an uncertainty metric. Here we used the uncorrelated standard deviation, as explained in the next sub-section.
The MSER defines the start of the equilibrated region \(\widehat{d}(n)\) by solving the minimization problem:
The enthalpy of adsorption was computed as
where N is the number of adsorbates on the simulation box and U is the potential energy64. All the adsorption enthalpy values are reported in kJ/mol and are the values as calculated by the pyMSER package. The Left-most Local Minimum (LLM) version of MSER was used in a batched data with batch size of 5.
Uncorrelated standard deviation (uSD)
To use an uncertainty metric that reflects, at the same time, the real dispersion of the simulated values and the number of cycles used for this simulation, the uncorrelated standard deviation (uSD) was used as an uncertainty metric. To calculate this quantity, first the number of uncorrelated states in the simulation is estimated by calculating the autocorrelation time. The autocorrelation time is estimated by calculating the autocorrelation function of the equilibrated data. An exponential decay function is fitted over the values of the autocorrelation function and the autocorrelation time is calculated as the half-life of this exponential decay.
The equilibrated data is then divided into chunks so that each chunk has a number of data points equivalent to the autocorrelation time. Then, the average value of each chunk is calculated and the standard deviation over this list of uncorrelated average values is calculated as the uSD. The uncorrelated standard deviation was obtained, as described above, using the pyMSER package.
Automatic simulation workflow
A set of scripts composed of three stages were created to automate the isotherm generation process. First, a pre-processing step is performed where partial charges are calculated for all structures. Next, a set of calculation scheduler scripts are executed, where steps such as copying the force field and CIF files, creating a supercell with P1 symmetry, writing the RASPA input file, running the RASPA simulation, parsing the RASPA output, and performing the MSER analysis of the results for averaging over the equilibrated phase of the simulation, are run in sequence.
Finally, a post-processing script is executed to analyze the results and resubmit incomplete calculations that exceeded the time allocated for the job submission queue. Whenever necessary, the simulations were resumed from a restart binary file generated by RASPA every 1000 cycles. A simplified scheme containing the main steps of this workflow is present on Fig. 1.
Revised autocorrelations (RACs) descriptors
To understand the diversity of our subset of CRAFTED structures, and understand how representative they are with respect to the MOF material class, revised autocorrelations (RACs) descriptors were calculated using the molSimplify software v1.7.165. RACs are built by generating a crystal graph derived from the adjacency matrix computed for the primitive cell of the crystal structure and calculating the discrete correlations between atomic properties (Pauling electronegativity, nuclear charge, etc.) over the atoms. The correlations are composed of the products (Equation 5) and the differences (Equation 6) of an atomic property P for atom i, which is selected from the start atom list and is correlated to atom j selected from the scope atom list when they are separated by d number of bonds.
Six atomic properties were used to compute RACs: atom identity (I), connectivity (T), Pauling electronegativity (χ), covalent radii (S), nuclear charge (Z) and polarisability (a). These properties are used to generate metal-centred, linker and functional-group descriptors. To generate a fixed length descriptor, the averages of these descriptors were used, thus generating 156 features (40 for metal chemistry, 68 for linker chemistry and 48 for functional group chemistry) for each MOF structure.
Dimensionality reduction and cluster analysis
To reduce the dimensionality of the feature vectors that describe CRAFTED structures, the t-Stochastic Neighbour Embedding (t-SNE)66 method was employed as implemented in the scikit-learn67 package v1.2.1. Different fitting parameters where used for each set of descriptors for metal chemistry (perplexity = 45, early_exaggeration = 1, learningrate = 50), linker chemistry (perplexity = 100, early_exaggeration = 1, learningrate = 50), functional groups chemistry (perplexity = 50, early_exaggeration = 1, learningrate = 200) and geometric properties (perplexity = 50, early_exaggeration = 1, learningrate = 200). The value 5792 was used as a random seed in all analyses.
For the t-SNE projections of the geometric properties, 14 features were used: largest included sphere (Dis), the largest free sphere (Dfs), largest included sphere along a free path (Disfs), volumetric accessible area (ASAm2/cm3), gravimetric accessible area (ASAm2/g), volumetric non-accessible area (NASAm2/cm3), gravimetric non-accessible area (NASAm2/g), unit cell volume, crystal density, accessible volume fraction (AVF), non-accessible volume fraction (NAVF), accessible volume (AVcm3/g), non-accessible volume (NAVcm3/g), and the number of pockets (npockets). All these properties were calculated using Zeo++ v0.368. The chemical descriptors used for the t-SNE projections of the MOF structures were described in the previous section. All structures that could not have their descriptors calculated were removed from the list.
For the unsupervised cluster analysis, we used the Density-Based Spatial Clustering of Applications with Noise (DBSCAN)69 method as implemented in scikit-learn. The DBSCAN analysis was performed using the standardized and scaled set of descriptors with eps = 0.09 and minimum number of samples per cluster of 25.
Data Records
The CRAFTED dataset59 is available in a dedicated Zenodo repository at https://doi.org/10.5281/zenodo.7689919. All the simulations were executed in a traditional HPC cluster with a heterogeneous assortment of Intel® Xeon® 2nd generation processors. Each simulated isotherm took on average ~11 CPU-hours to compute, which translates to ~62 CPU-years to generate the full dataset.
CRAFTED provides 49,680 isotherm files and 49,680 adsorption enthalpy files resulting from the GCMC simulation of two gases (CO2 and N2) on 690 MOF structures at three temperatures (273, 298, and 323 K) using two force fields (UFF and DREIDING) and six partial charge methods (no charges, Qeq, EQeq, PACMOF, MPNN, and DDEC). Alongside the isotherm data, the charge-assigned CIF files, force field and molecule definition files are provided, to ensure reproducibility and facilitate a future database expansion.
Each isotherm file corresponds to a comma-separated value (CSV) file containing three labeled columns corresponding to pressure (in Pa), uptake volume and its uncertainty (in mol/kg). The file names follow the pattern Q_MOF_FF_GAS_T.csv, therefore the isotherm file named DDEC_ABUWOJ_UFF_CO2_273.csv contains the data corresponding to the CO2 adsorption isotherm at 273 K on the ABUWOJ MOF with DDEC partial charges using the UFF force field. The adsorption enthalpy file names follows the same pattern.
The adsorption enthalpy files correspond to a CSV file containing three labeled columns corresponding to pressure (in Pa), adsorption enthalpy and its uncertainty (in kJ/mol), following the same naming pattern as the isotherm files.
The RASPA input file names (Q_MOF_FF_GAS_T.input) follows the same pattern as the isotherm files, the CIF files are separated into folders according to their partial charge (Qeq, EQeq, DDEC, PACMOF, MPNN and NEUTRAL) type and the force field files are separated into folder according to their type (UFF and DREIDING).
Technical Validation
Chemical and geometrical diversity of CRAFTED structures
The selection of a subset of structures that can be modeled simultaneously by both UFF and DREIDING force fields may impose a limitation on the structural and chemical representativeness of the CRAFTED database and, consequently, on the results obtained with this data. To ensure that CRAFTED structures form a group that represents the great diversity of experimentally realised MOFs, both the geometrical and chemical diversities must be represented.
To evaluate the geometrical diversity, the pore size (such as the largest included sphere, largest free sphere and largest included sphere along a free path), void fraction, density, unit cell volume, specific area (both gravimetric and volumetric), pore volume (both gravimetric and volumetric) and the number of pockets (non accessible pores) were used as the descriptors for t-SNE projection. Both the accessible and non-accessible specific area and pore volume was used.
For the chemical diversity, the revised autocorrelations (RACs) descriptors70 were used. This approach has been successfully applied to study transition metal chemistry71 and the chemical diversity of MOF datasets72. The chemical characteristics of the MOFs were divided into three categories: metal node chemistry, organic linker chemistry and functional groups chemistry.
Figure 2 shows the t-SNE projection onto 2D maps of the four selected groups of descriptors for the structures in the CoRE MOF 2019 database20,21 (colored circles) and the selected structures for CRAFTED database (red hexagons). Although CRAFTED structures were taken from the first version of CoRE MOF, from 2014, here the comparison is made with the second version of this database, from 2019, as it has a greater number and diversity of structures.
t-SNE representation of the CRAFTED structures (red points) and the CoRE MOF 201920,21 (coloured points) database on different domains of MOF chemistry based on RACs descriptors and geometric properties. (a) metal chemistry, (b) linker chemistry, (c) functional groups chemistry, (d) geometric properties. The color scheme correspond to the cluster assigned by DBSCAN.
To numerically evaluate the overlap of the databases in t-SNE projected space, the DBSCAN method was used. This method is an unsupervised machine learning technique used to identify clusters of varying shape and size, grouping points that are close to each other. Since the t-SNE method reduces the feature space by modelling structures with similar features as nearby points and distinct features as distant points, the groups found by the DBSCAN method will share similarities within the original feature space.
The limitation imposed by the DREIDING force field reduces the diversity of metal chemistry observed in the CRAFTED database compared to the structures presented in CoRE MOF 2019, as shown in Fig. 2a. However, 55 of the 95 clusters found (58%) have some structure present in CRAFTED, indicating that even with a limited metal cluster composition, the CRAFTED structures show a good representation of the chemical diversity present in CoRE MOF 2019.
The chemical diversity of both organic linker and functional groups is much better represented within the CRAFTED structures, as can be seen in Fig. 2b,c. In both cases, one can see that the points from both CRAFTED and CoRE MOF 2019 structures are equally dispersed across 2D space. Additionally, 32 of the 35 clusters identified for linker chemistry (91%) and 40 of the 43 clusters identified for functional group chemistry (93%) contain structures present in CRAFTED.
The geometric properties are also well represented by the CRAFTED database, as shown in Fig. 2d. From the 27 clusters identified, 20 (74%) present structures from CRAFTED. Additionally, Fig. 3 shows the distribution density of the values for the main geometric properties presented by the structures on CRAFTED and CoRE MOF 2019. One sees that both databases contain similar distributions, showing that even with a smaller number of structures, the CRAFTED database is exemplary of synthesized MOF structural properties.
General impact of force field and partial charge selection
To illustrate the impact of molecular-level simulation parameters (force field and partial charge) on the outcome of the GCMC simulations, we show in Fig. 4 the absolute uptake and enthalpy of adsorption of CO2 on the MULQOA MOF at 237 K. At 0.1 bar, a typical pressure used in adsorption-based pressure swing adsorption (PSA) processes for CO2 capture73, the uptake values range from 0.5 to 5.0 mol/kg and every combination of partial charge and force field yields a different value, while at 10 bar almost all conditions resulted in similar uptake. This dispersion of results is also reflected on the enthalpy of adsorption that ranges from −17 to −45 kJ/mol and are fairly different for every combination of parameters.
Among the CRAFTED materials, one finds a diversity of responses to the choices of force field and partial charge method. Four representative cases are highlighted in Fig. 5. For some materials, the uptake is highly dependent on both the force field and partial charge, others are only sensitive to one of these parameters, and some are not sensitive to any. Therefore it is possible to anticipate that most studies that depend directly on the results of these molecular simulations, such as high-throughput computational screening or multiscale processes modelling, may also present different degrees of dependence on the choice of parameters.
Examples of four representative behaviours found in the dataset: (a) high sensitivity to force field and partial charge, (b) high sensitivity to force field and low sensitivity to partial charge, (c) low sensitivity to force field and high sensitivity to partial charge, and (d) low sensitivity to force field and partial charge.
Usage Notes
The CRAFTED database contains 49,680 CSV files with the isotherm adsorption data (pressure, uptake, and uncertainty), definition files for UFF and DREIDING force fields, and a total of 4,140 CIF files combining 690 structures with six partial charges (NEUTRAL, DDEC, PACMOF, MPNN, EQeq, and Qeq). The database also contains 49,680 files containing the adsorption enthalpy, and two CSV files containing the set of RAC and geometrical descriptors calculated with molSimplify and Zeo++, respectively. The 49,680 RASPA input files are provided to facilitate the reproduction of the isotherm simulations.
To facilitate the exploration and visualization of the isotherms and enthalpies of adsorption present on CRAFTED, we also developed an interactive visualization interface based on Jupyter notebooks and panel (v0.14.3). This interface allows the user to select a set of conditions–e.g. partial charge, temperature, force field, adsorbed gas, and material name–for each isotherm, thus facilitating a quick and easy visual comparison of the data. In addition, it is possible to download the CIF files, the inputs for the GCMC simulation with RASPA, and the data of the selected isotherms, thus facilitating the reproduction of the results present in CRAFTED. An example of the interface is shown in Fig. 6. We recommend the user to set up a Python environment using the environment.yml file provided therein.
A Jupyter notebook with the code to perform the t-SNE dimensionality reduction and the DBSCAN unsupervised clusterization analysis with the necessary files containing the RAC and geometric descriptors for the CoRE MOF 2019 is also provided alongside the CRAFTED data, providing an easy reproduction of the results presented in Fig. 2. Please note that, despite having frozen the t-SNE input values, random number generator seed and dependency versions, we observe minor discrepancies in the results when running on different CPU architectures, but which do not alter their interpretation. The results presented herein refer to Intel® x86_64 running Fedora Workstation 37.
To extend the application of the data present on CRAFTED, we prepared a Jupyter notebook with codes that apply the Ideal Adsorbed Solution Theory (IAST) to estimate the CO2/N2 mixture adsorption uptake and selectivity. We leveraged the pyGAPS package74 (v4.4.2) to model the pure component isotherms and predict the multi-component mixture isotherms.
Finally, we would like to highlight some points to show why this database benefits the scientific community. Machine learning (ML) and data-driven methods have become useful tools to aid in the discovery of new materials for CO2 capture. For example, surrogate models can be constructed from GCMC-simulated adsorption data to map the structure-property relationship of nanoporous materials, which may then be used to accelerate the HTCS of previously unexplored databases of adsorbents75,76. Deep generative models can be trained with simulated adsorption data to develop property-orientated generative algorithms to discover new materials on the latent chemical space optimized for gas capture applications by an inverse design approach77,78,79.
As ML model prediction accuracy and data quality are intrinsically related, there is an apparent requirement to assess the impact of uncertainty from molecular-level simulations on the confidence of machine learning model predictions80. The efficacy of the surrogate models, for example, depends on the quality of information provided by the material feature vector, and so concerted efforts have been made to develop useful representations of MOFs81. However, little is known about the impact of force field selection on the interpretability of surrogate model predictions. Particularly in the case of MOFs with coordinatively unsaturated metals, different generic force fields and charge assignment schemes can deliver dramatically different results.
Therefore, the importance of material features–learned either explicitly by the surrogate model as in decision trees, or extracted through feature permutations/SHAP values82–may be subject to considerable discrepancies. Feature importance analyses are useful to guide the design of new functional MOFs, and so it is desirable to understand the differences (if any) that arise from different levels of molecular theory.
Code availability
The Jupyter notebooks providing the panel visualisation of the isotherm curves, enthalpy of adsorption data, IAST-based multicomponent mixture isotherm, and the t-SNE + DBSCAN analysis of the chemical and geometric properties of MOFs are distributed alongside the database in the Zenodo59 repository. A fully automated workflow that is capable of recreating the dataset was made available as an open-source project (v1.0.0) on GitHub (https://github.com/st4sd/nanopore-adsorption-experiment).
References
Mac Dowell, N., Fennell, P. S., Shah, N. & Maitland, G. C. The role of CO2 capture and utilization in mitigating climate change. Nature Climate Change 7, 243–249 (2017).
Metz, B., Davidson, O., De Coninck, H., Loos, M. & Meyer, L. IPCC special report on carbon dioxide capture and storage (Cambridge: Cambridge University Press, 2005).
Sholl, D. S. & Lively, R. P. Seven chemical separations to change the world. Nature 532, 435–437 (2016).
Samanta, A., Zhao, A., Shimizu, G. K., Sarkar, P. & Gupta, R. Post-combustion CO2 capture using solid sorbents: a review. Industrial & Engineering Chemistry Research 51, 1438–1463 (2012).
Yaghi, O. M. et al. Reticular synthesis and the design of new materials. Nature 423, 705–714 (2003).
Furukawa, H., Cordova, K. E., O’Keeffe, M. & Yaghi, O. M. The chemistry and applications of metal-organic frameworks. Science 341, 1230444 (2013).
Chen, Z., Kirlikovali, K. O., Li, P. & Farha, O. K. Reticular chemistry for highly porous metal–organic frameworks: The chemistry and applications. Accounts of Chemical Research 55, 579–591 (2022).
Maia, R. A., Louis, B., Gao, W. & Wang, Q. CO2 adsorption mechanisms on MOFs: a case study of open metal sites, ultra-microporosity and flexible framework. Reaction Chemistry & Engineering 6, 1118–1133 (2021).
Cote, A. P. et al. Porous, crystalline, covalent organic frameworks. Science 310, 1166–1170 (2005).
Diercks, C. S. & Yaghi, O. M. The atom, the molecule, and the covalent organic framework. Science 355, eaal1585 (2017).
Maia, R. A. et al. CO2 Capture by Hydroxylated Azine-Based Covalent Organic Frameworks. Chemistry–A European Journal 27, 8048–8055 (2021).
Banerjee, R. et al. High-throughput synthesis of zeolitic imidazolate frameworks and application to CO2 capture. Science 319, 939–943 (2008).
Yang, J. et al. Principles of designing extra-large pore openings and cages in zeolitic imidazolate frameworks. Journal of the American Chemical Society 139, 6448–6455 (2017).
Wang, H., Pei, X., Kalmutzki, M. J., Yang, J. & Yaghi, O. M. Large cages of zeolitic imidazolate frameworks. Accounts of Chemical Research 55, 707–721 (2022).
Lin, L.-C. et al. In silico screening of carbon-capture materials. Nature Materials 11, 633–641 (2012).
Chung, Y. G. et al. Computation-ready, experimental metal–organic frameworks: A tool to enable high-throughput screening of nanoporous crystals. Chemistry of Materials 26, 6185–6192 (2014).
Chung, Y. G. et al. Computation-Ready, Experimental Metal–Organic Frameworks. Zenodo https://doi.org/10.5281/zenodo.3228673 (2014).
Tong, M., Lan, Y., Yang, Q. & Zhong, C. Exploring the structure-property relationships of covalent organic frameworks for noble gas separations. Chemical Engineering Science 168, 456–464 (2017).
Moghadam, P. Z. et al. Development of a cambridge structural database subset: a collection of metal–organic frameworks for past, present, and future. Chemistry of Materials 29, 2618–2625 (2017).
Chung, Y. G. et al. Advances, updates, and analytics for the computation-ready, experimental metal–organic framework database: CoRE MOF 2019. Journal of Chemical & Engineering Data 64, 5985–5998 (2019).
Chung, Y. G. et al. Computation-Ready Experimental Metal-Organic Framework (CoRE MOF) 2019 Dataset. Zenodo https://doi.org/10.5281/zenodo.3528250 (2019).
Ongari, D., Yakutovich, A. V., Talirz, L. & Smit, B. Building a consistent and reproducible database for adsorption evaluation in covalent–organic frameworks. ACS Central Science 5, 1663–1675 (2019).
Colón, Y. J., Gomez-Gualdron, D. A. & Snurr, R. Q. Topologically guided, automated construction of metal–organic frameworks and their evaluation for energy-related applications. Crystal Growth & Design 17, 5801–5810 (2017).
Rosen, A. S. et al. Machine learning the quantum-chemical properties of metal–organic frameworks for accelerated materials discovery. Matter 4, 1578–1597 (2021).
Lyu, H., Ji, Z., Wuttke, S. & Yaghi, O. M. Digital reticular chemistry. Chem 6, 2219–2241 (2020).
Ji, Z. et al. From molecules to frameworks to superframework crystals. Advanced Materials 33, 2103808 (2021).
Colón, Y. J. & Snurr, R. Q. High-throughput computational screening of metal–organic frameworks. Chemical Society Reviews 43, 5735–5749 (2014).
Majumdar, S., Moosavi, S. M., Jablonka, K. M., Ongari, D. & Smit, B. Diversifying databases of metal organic frameworks for high-throughput computational screening. ACS Applied Materials & Interfaces 13, 61004–61014 (2021).
Dubbeldam, D., Walton, K. S., Vlugt, T. J. & Calero, S. Design, parameterization, and implementation of atomic force fields for adsorption in nanoporous materials. Advanced Theory and Simulations 2, 1900135 (2019).
Rappé, A. K., Casewit, C. J., Colwell, K., Goddard, W. A. III & Skiff, W. M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. Journal of the American Chemical Society 114, 10024–10035 (1992).
Mayo, S. L., Olafson, B. D. & Goddard, W. A. Dreiding: a generic force field for molecular simulations. Journal of Physical Chemistry 94, 8897–8909 (1990).
Potoff, J. J. & Siepmann, J. I. Vapor–liquid equilibria of mixtures containing alkanes, carbon dioxide, and nitrogen. AIChE Journal 47, 1676–1682 (2001).
Allen, M. P. & Tildesley, D. J. Computer simulation of liquids (Oxford University Press, 2017).
Jorgensen, W. L. Optimized intermolecular potential functions for liquid alcohols. The Journal of Physical Chemistry 90, 1276–1284 (1986).
Bayly, C. I., Cieplak, P., Cornell, W. & Kollman, P. A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the resp model. The Journal of Physical Chemistry 97, 10269–10280 (1993).
Breneman, C. M. & Wiberg, K. B. Determining atom-centered monopoles from molecular electrostatic potentials. the need for high sampling density in formamide conformational analysis. Journal of Computational Chemistry 11, 361–373 (1990).
Campañá, C., Mussard, B. & Woo, T. K. Electrostatic potential derived atomic charges for periodic systems using a modified error functional. Journal of Chemical Theory and Computation 5, 2866–2878 (2009).
Manz, T. A. & Limas, N. G. Introducing DDEC6 atomic population analysis: part 1. Charge partitioning theory and methodology. RSC Advances 6, 47771–47801 (2016).
Limas, N. G. & Manz, T. A. Introducing DDEC6 atomic population analysis: part 2. Computed results for a wide range of periodic and nonperiodic materials. RSC Advances 6, 45727–45747 (2016).
Manz, T. A. Introducing DDEC6 atomic population analysis: part 3. Comprehensive method to compute bond orders. RSC Advances 7, 45552–45581 (2017).
Rappe, A. K. & Goddard III, W. A. Charge equilibration for molecular dynamics simulations. The Journal of Physical Chemistry 95, 3358–3363 (1991).
Ramachandran, S., Lenz, T., Skiff, W. & Rappé, A. Toward an understanding of zeolite Y as a cracking catalyst with the use of periodic charge equilibration. The Journal of Physical Chemistry 100, 5898–5907 (1996).
Wilmer, C. E., Kim, K. C. & Snurr, R. Q. An extended charge equilibration method. The Journal of Physical Chemistry Letters 3, 2506–2511 (2012).
Wells, B. A., De Bruin-Dickason, C. & Chaffee, A. L. Charge equilibration based on atomic ionization in metal–organic frameworks. The Journal of Physical Chemistry C 119, 456–466 (2015).
Kancharlapalli, S., Gopalan, A., Haranczyk, M. & Snurr, R. Q. Fast and accurate machine learning strategy for calculating partial atomic charges in metal–organic frameworks. Journal of Chemical Theory and Computation 17, 3052–3064 (2021).
Raza, A., Sturluson, A., Simon, C. M. & Fern, X. Message passing neural networks for partial charge assignment to metal–organic frameworks. The Journal of Physical Chemistry C 124, 19070–19082 (2020).
Zheng, C., Liu, D., Yang, Q., Zhong, C. & Mi, J. Computational study on the influences of framework charges on CO2 uptake in metal-organic frameworks. Industrial & Engineering Chemistry Research 48, 10479–10484 (2009).
Hamad, S., Balestra, S. R., Bueno-Perez, R., Calero, S. & Ruiz-Salvador, A. R. Atomic charges for modeling metal–organic frameworks: Why and how. Journal of Solid State Chemistry 223, 144–151 (2015).
Ongari, D. et al. Evaluating charge equilibration methods to generate electrostatic fields in nanoporous materials. Journal of Chemical Theory and Computation 15, 382–401 (2018).
Sladekova, K. et al. The effect of atomic point charges on adsorption isotherms of CO2 and water in metal organic frameworks. Adsorption 26, 663–685 (2020).
Altintas, C. & Keskin, S. Role of partial charge assignment methods in high-throughput screening of MOF adsorbents and membranes for CO2/CH4 separation. Molecular Systems Design & Engineering 5, 532–543 (2020).
Liu, S. & Luan, B. Benchmarking various types of partial atomic charges for classical all-atom simulations of metal-organic frameworks. Nanoscale (2022).
Li, S., Chung, Y. G. & Snurr, R. Q. High-throughput screening of metal–organic frameworks for CO2 capture in the presence of water. Langmuir 32, 10368–10376 (2016).
Boyd, P. G. et al. Data-driven design of metal–organic frameworks for wet flue gas CO2 capture. Nature 576, 253–256 (2019).
Avci, G., Erucar, I. & Keskin, S. Do new MOFs perform better for CO2 capture and H2 purification? Computational screening of the updated MOF database. ACS Applied Materials & Interfaces 12, 41567–41579 (2020).
Deng, X. et al. Large-scale screening and machine learning to predict the computation-ready, experimental metal-organic frameworks for CO2 capture from air. Applied Sciences 10, 569 (2020).
Yan, Y. et al. Machine learning and in-silico screening of metal–organic frameworks for O2/N2 dynamic adsorption and separation. Chemical Engineering Journal 427, 131604 (2022).
Gasteiger, J. & Marsili, M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36, 3219–3228 (1980).
Oliveira, F. L. et al. CRAFTED - An exploratory database of simulated adsorption isotherms of metal-organic frameworks. Zenodo https://doi.org/10.5281/zenodo.7689919 (2023).
Dubbeldam, D., Calero, S., Ellis, D. E. & Snurr, R. Q. RASPA: molecular simulation software for adsorption and diffusion in flexible nanoporous materials. Molecular Simulation 42, 81–101 (2016).
Dubbeldam, D., Torres-Knoop, A. & Walton, K. S. On the inner workings of monte carlo codes. Molecular Simulation 39, 1253–1292 (2013).
Peng, D. & Robinson, D. A new two-constant equation of state. Industrial Engineering Chemistry Fundamentals 15, 59–64 (1976).
White, K. P. Jr An effective truncation heuristic for bias reduction in simulation output. Simulation 69, 323–334 (1997).
Vlugt, T., Garca-Pérez, E., Dubbeldam, D., Ban, S. & Calero, S. Computing the heat of adsorption using molecular simulations: the effect of strong Coulombic interactions. Journal of Chemical Theory and Computation 4, 1107–1118 (2008).
Ioannidis, E. I., Gani, T. Z. H. & Kulik, H. J. molsimplify: A toolkit for automating discovery in inorganic chemistry. Journal of Computational Chemistry 37, 2106–2117 (2016).
Van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. Journal of Machine Learning Research 9 (2008).
Pedregosa, F. et al. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research 12, 2825–2830 (2011).
Willems, T. F., Rycroft, C. H., Kazi, M., Meza, J. C. & Haranczyk, M. Algorithms and tools for high-throughput geometry-based analysis of crystalline porous materials. Microporous and Mesoporous Materials 149, 134–141 (2012).
Bi, F., Wang, W. & Chen, L. DBSCAN: density-based spatial clustering of applications with noise. J. Nanjing Univ 48, 491–498 (2012).
Janet, J. P. & Kulik, H. J. Resolving transition metal chemical space: Feature selection for machine learning and structure–property relationships. The Journal of Physical Chemistry A 121, 8939–8954 (2017).
Nandy, A., Duan, C., Janet, J. P., Gugler, S. & Kulik, H. J. Strategies and software for machine learning accelerated discovery in transition metal chemistry. Industrial & Engineering Chemistry Research 57, 13973–13986 (2018).
Moosavi, S. M. et al. Understanding the diversity of the metal-organic framework ecosystem. Nature Communications 11, 1–10 (2020).
Park, J. et al. How well do approximate models of adsorption-based CO2 capture processes predict results of detailed process models? Industrial & Engineering Chemistry Research 59, 7097–7108 (2019).
Iacomi, P. & Llewellyn, P. L. pyGAPS: a Python-based framework for adsorption isotherm processing and material characterisation. Adsorption 25, 1533–1542 (2019).
Simon, C. M., Mercado, R., Schnell, S. K., Smit, B. & Haranczyk, M. What are the best materials to separate a xenon/krypton mixture? Chemistry of Materials 27, 4459–4475 (2015).
Bucior, B. J. et al. Energy-based descriptors to rapidly predict hydrogen storage in metal–organic frameworks. Molecular Systems Design & Engineering 4, 162–174 (2019).
Sanchez-Lengeling, B. & Aspuru-Guzik, A. Inverse molecular design using machine learning: Generative models for matter engineering. Science 361, 360–365 (2018).
Yao, Z. et al. Inverse design of nanoporous crystalline reticular materials with deep generative models. Nature Machine Intelligence 3, 76–86 (2021).
Pollice, R. et al. Data-driven strategies for accelerated materials design. Accounts of Chemical Research 54, 849–860 (2021).
Nigam, A. et al. Assigning confidence to molecular property prediction. Expert Opinion on Drug Discovery 16, 1009–1023 (2021).
Jablonka, K. M., Ongari, D., Moosavi, S. M. & Smit, B. Big-data science in porous materials: materials genomics and machine learning. Chemical Reviews 120, 8066–8129 (2020).
Lundberg, S. M., Erion, G. G. & Lee, S.-I. Consistent individualized feature attribution for tree ensembles. Preprint at: https://arxiv.org/abs/1802.03888 (2018).
Acknowledgements
The authors would like to acknowledge Flor Siperstein and Joseph Manning (University of Manchester) for fruitful discussions that helped shape this work. The authors would also like to acknowledge Manuela Rodriguez, Alessandro Pomponio and Vassilis Vassiliadis (IBM Research) for their support in the open-source release of the simulation code.
Author information
Authors and Affiliations
Contributions
F.L.O. developed the GCMC simulation workflow, analyzed the data, compiled the database and wrote the manuscript. C.C. wrote the manuscript and proposed the generation of the database. R.N.B.F. developed the GCMC simulation workflow and wrote the manuscript. B.L. developed the GCMC simulation workflow. A.H.F. proposed the generation of the database. L.S. and M.S. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oliveira, F.L., Cleeton, C., Neumann Barros Ferreira, R. et al. CRAFTED: An exploratory database of simulated adsorption isotherms of metal-organic frameworks. Sci Data 10, 230 (2023). https://doi.org/10.1038/s41597-023-02116-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02116-z