Abstract
Multi-organ dysfunction is a major complication after spinal cord injury (SCI). In addition to local injury within the spinal cord, SCI causes major disruption to the peripheral organ innervation and regulation. The liver contains sympathetic, parasympathetic, and small sensory axons. The bi-directional signaling of sensory dorsal root ganglion (DRG) neurons that provide both efferent and afferent information is of key importance as it allows sensory neurons and peripheral organs to affect each other. SCI-induced liver inflammation precedes and may exacerbate intraspinal inflammation and pathology after SCI, which may be modulated by activity and exercise. In this study, we collected comprehensive gene expression data through RNA sequencing of liver tissue from rats with chronic SCI to determine the effects of activity and exercise on those expression patterns. The sequenced data are of high quality and show a high alignment rate to the Rn6 genome. Gene expression is demonstrated for genes associated with known liver pathologies. UCSC Genome Browser expression tracks are provided with the data to facilitate exploration of the samples.
Design Type(s) | gene expression analysis objective • stimulus or stress design • transcription profiling design |
Measurement Type(s) | gene expression |
Technology Type(s) | RNA sequencing |
Factor Type(s) | Spinal Cord Injury • Exercise • EnvironmentalVariable • biological replicate |
Sample Characteristic(s) | Rattus norvegicus • liver |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
More than 10,000 people in the U.S. suffer from traumatic spinal cord injury (SCI) yearly, and the number of people living with paralysis is ~5.4 million1. Beyond impairments to sensation and voluntary movement, SCI disturbs the sensory and autonomic nervous systems (ANS) and induces dysfunction in multiple organ systems2. Neurogenic pain, depression, cardiovascular disease, liver damage, kidney dysfunction and urinary tract infection are all common in SCI patients and hinder functional recovery and affect long term quality of life. These SCI-induced changes, compounded by a sedentary lifestyle, lead to obesity and a host of metabolic changes, with ~55% of SCI individuals developing frank metabolic syndrome3. SCI also triggers a systemic inflammatory response (SIR) that contributes to a high incidence of secondary organ complications4,5,6,7. Several studies have suggested that post-SCI, the liver contributes to the initiation and propagation of the SIR4,7,8. Rodent studies show that traumatic SCI triggers neutrophil infiltration, macrophage activation, and expression of pro-inflammatory cytokines and chemokines in the liver which amplifies the central nervous system (CNS) response to injury4. Specifically, liver Kupffer cells control the magnitude of the inflammatory response in the injured brain and spinal cord8. This inflammation in the liver appears as early as 30 minutes after injury9 and its severity is correlated with lesion level10. Further, various liver abnormalities have been observed chronically in humans and rodents post-SCI including non-alcoholic fatty liver disease (humans)11 as well as considerable hepatic lipid accumulation (rodents)12. These liver pathologies are sustained chronically post-SCI.
Several clinical studies suggest that physical activity can be effective in enhancing recovery of function and also in ameliorating SCI-induced cardiometabolic syndrome, risks of dyslipidemia, and insulin resistance in humans13,14,15,16. However the underlying mechanisms responsible for these outcomes largely remain unknown. Previous studies have suggested that spontaneous improvements in locomotor function are related to “in-cage activity”17,18,19,20, indicating that this activity acts as a type of ‘rehabilitative therapy’. This is important because human SCI patients are highly restricted in their activity levels post-SCI. The main objective of this study is to examine the chronic transcriptomic changes in rat liver after SCI and evaluate how this profile is modified with spontaneous in-cage activity and specific forms of exercise, with the premise that these changes may suggest mechanisms.
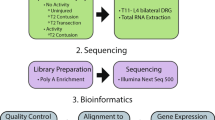
An overall workflow of the study is schematically represented in Fig. 1 with our experimental design given in greater detail in Fig. 2. For this initial look at the effect of activity/exercise on SCI, we chose female adult rats (~8–9 weeks old) to control for transcriptomic responses related to age and gender. In our experience, female rats recover more quickly from surgery and have greater motivation to exercise, providing our best chance for identifying transcriptomic changes related to activity/exercise. We used two approaches to explore the effect of activity and exercise on injury. In our initial approach, referred to as SCI + In-Cage Activity, we obtained liver samples from contusion (CONT) injured rats housed in large (activity-enhanced) or tiny (activity-restricted) cages. In the latter, in-cage activity is reduced by 75–80% versus large cages for both SCI and naïve groups (DSKM, unpublished observations). The tiny, activity-restricted cages mimic the clinical situation in which sedentary behaviors are enhanced, thereby contributing to the general decline in physical health. In our second approach, referred to as SCI + Exercise, we obtained liver samples from rats housed in activity-restricted (tiny) cages that received one of two different exercise training paradigms: swimming (SWIM) or shallow water walking (SWW), each with its own range of beneficial effects in terms of recovery of function19,20,21,22. In addition, another group of animals received a complete spinal cord transection (TX) to examine the transcriptome in the total absence of descending, supraspinal innervation.
Experimental workflow. (1) In the SCI + In-Cage Activity sample set (top left), rats with CONT injury were housed in large or tiny cages. Rats with no SCI were housed in standard size cages. In the SCI + Exercise sample set (top right), rats received a CONT SCI, a TX SCI, or no SCI. A subset of rats with CONT SCI were exposed to swimming (SWIM) or shallow water walking (SWW) after injury. For both sample sets, RNA was extracted from the liver. (2) RNA samples were subjected to poly A enrichment and sequenced on an Illumina NextSeq 500. (3) The bioinformatics workflow included quality control analysis, alignment to the Rattus norvegicus (Rn6) reference assembly, and sample expression analysis with UCSC Genome Browser visualization.
A quality control analysis of our data indicates high quality sequenced reads with high alignment rates to the Rattus norvegicus genome (Rn6). Gene expression data indicate high levels of gene activity from categories relevant to SCI-induced liver pathologies (see Fig. 3 for mean expression in CONT injury and No SCI). To facilitate exploration of expression across samples, UCSC Genome Browser23 tracks were created and made available with this dataset. In Fig. 4, expression for two enzymes with a specific role in lipid metabolism, apolipoprotein A1 (Apoa1) (Fig. 4a) and cytochrome P450 1A2 (Cyp1a2) (Fig. 4b) is displayed for CONT SCI samples. The raw data presented here are offered as a valuable resource to the scientific community for future investigation and further elucidation of underlying biological pathways related to SCI-induced pathogenicity in the liver.
Gene expression in categories relevant to SCI-induced liver pathologies. Five genes associated with four known liver pathologies with the highest mean expression in liver from animals with CONT SCI (with activity restriction by housing in tiny cages) are presented (taken from the SCI + Exercise set). Mean expression for No SCI is included as a comparison. Read counts are normalized using DESeq2’s default method, relative log expression (RLE).
Methods
Animals
All animal procedures were performed in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Research Council, 1996) and the University of Louisville Institutional Animal Care and Use Committee. Female Sprague Dawley rats of body weight 235–249 g were obtained from Sprague Dawley, Inc. (Indianapolis, IN). All rats were initially housed in standard cages and maintained in a 12h-light/dark cycle throughout. Tap water and a standard rodent diet were available to all rats ad libitum.
Experimental design and SCI
The experimental workflow from sample collection through sequencing and bioinformatics is displayed in Fig. 1. The experimental design is illustrated in Fig. 2. Prior to the study, 15 animals in the SCI + In-Cage Activity study were randomly divided into three groups: no injury housed in standard cages (No SCI + Standard, 5 replicates), T2 contusion injury housed in tiny cages (CONT SCI + Tiny, 5 replicates), and T2 contusion injury housed in large cages (CONT SCI + Large, 5 replicates). In the SCI + Exercise study, 22 animals were randomly assigned to five groups: no injury (No SCI, 4 replicates), T2 transection injury (TX SCI, 4 replicates), T2 contusion injury (CONT SCI, 4 replicates), T2 contusion injury followed by swimming as exercise (CONT SCI + SWIM, 5 replicates), and T2 contusion injury followed by shallow water walking as exercise (CONT SCI + SWW, 5 replicates). Throughout the study, rats were doubly-housed with individuals from the same experimental group.
All rats were initially gentled for two weeks, during which time they were introduced and acclimated to the testing and exercise facilities. After this period, animals were anaesthetized with a ketamine (50 mg/kg)/xylazine (0.024 mg/kg)/acepromazine (0.005 mg/kg) cocktail (IP) and given glycopyrolate (0.08 mg/kg, IM) prior to SCI surgeries. For all injury groups (CONT and TX), a dorsal midline incision was made in the superficial muscle overlying the T1–T3 vertebrae. A single level laminectomy was then performed at the T2 vertebrae. Animals in the CONT groups received a moderately severe contusion injury (25 g-cm SCI) at the T2 spinal cord level using the NYU Impactor24,25. For animals in the TX group, a scalpel was used to deliver a complete transection of the spinal cord at T2. The muscle and skin overlying the injury were sutured in layers and antibiotic ointment was applied to the incision. Injured animals were monitored on heating pads until they recovered from the anesthesia. Rats were then doubly-housed in cages with ALPHA-dri® bedding (Shepherd’sTM Specialty Paper, Milford, NJ) for the remainder of the study. Post-operative care consisted of daily injections of gentamicin sulfate for 7 days (20 mg/kg, S.C.), twice-daily injections of buprenorphine for 3 days (0.03 mg/kg, S.C., and as needed for pain management thereafter), and twice-daily 5 ml boluses of lactated ringers for three days (and as needed for hydration thereafter). Manual bladder expression was conducted three times a day until reflexive voiding was re-established. Rats were maintained on a 12-hour day/night light cycle throughout and had access to standard rat chow and water ad libitum. During the 2 week gentling and a 3 day recovery period, all animals were doubly housed in standard cages, measuring 22″ × 12.5″ × 8″.
Three days after injury, animals in the SCI + In-Cage Activity study were doubly housed in tiny cages (7.5″ × 8.5″ × 8″) to restrict movement and activity or large cages (14″ × 18″ × 8″, base dimension; 16″ × 20″ ceiling) to allow for greater movement for the duration of the study. No SCI controls remained in standard cages.
Three days after injury, all animals in the SCI + Exercise study were doubly-housed in tiny cages to restrict in-cage activity for the duration of the study. Animals in the CONT SCI + SWIM and CONT SCI + SWW groups began exercising 14 days post-injury. Exercise sessions were conducted 5 consecutive days/week for 10 weeks. Animals exercised for 30 minutes each day with 15 minutes of exercise in the morning and 15 minutes in the afternoon. The morning and afternoon sessions were separated by a minimum of one hour. Each 15 minute session consisted of three five minute periods of exercise with breaks between the periods lasting approximately 20–25 minutes.
Tissue collection and RNA extraction
Animals were sacrificed with a ketamine overdose at 8.5, 11.5, or 13.5 weeks post-SCI, depending on condition (see Fig. 2). All uninjured animals were sacrificed at a time point equivalent to 11.5 weeks post-SCI in the other animals. Hearts were arrested in diastole with an injection of 3 M KCl. Animals were perfused with PBS supplemented with 20% RNA later (Ambion, Life Technologies, Carlsbad, CA). Livers were taken from each animal and 200 mg of liver tissue was processed from each using RNeasy Lipid Tissue Mini Kit (Qiagen) to isolate RNA.
Library preparation and sequencing
1 µg of total RNA samples were used for poly A enrichment. First and second strands were synthesised followed by 3′ end adenylation. Samples were barcoded with Illumina TrueSeq adapters. 1.8 pM of barcoded library was denatured, and sequencing was performed on the University of Louisville Center for Genetics and Molecular Medicine (CGeMM) Illumina NextSeq 500 using the NextSeq 500/550 1X75 cycle High output kit (Illumina, Carlsbad, CA).
RNA-seq data analysis
Sequencing produced over 1 billion single end reads across the 37 samples. The vast majority of read lengths fell between 74–76 bases across all samples. The quality of the reads was assessed using FastQC v.0.10.126, which indicated no sequence trimming was necessary. The sequences were directly aligned to the Rattus norvegicus reference genome assembly (Rn6) using Star version 2.627. Read counts for gene regions were obtained with HTSeq (version 0.10.0)28 using Ensembl annotations29 (Rn6 version 93). The annotation file was parsed to exclude mitochondria genes in an effort to reduce non-relevant variation in subsequent steps of the analysis. The resulting annotation file extracted read counts for 24,613 gene locations.
A principal component analysis (PCA) was performed using the R programming language30 to examine within- and between-group variability of the samples. Three-dimensional PCA plots were generated using the R package ‘pca3d’. DESeq2’s regularized log transformation31,32 was applied to the raw counts prior to PCA to reduce the effect of high level variation genes on the spread of sample points.
Prior to examining gene expression, raw read counts were normalized to remove natural variation across samples arising from differences in tissue sampling and sequencing using DESeq2’s default method, relative log expression (RLE)31,32,33. UCSC Genome Browser tracks were created to facilitate exploration of gene expression in each of the samples23. The tracks were created using methods and available utilities described on the UCSC Genome Browser website for converting sequencing alignment files in BAM format to BigWig format.
Data Records
The data were submitted to NCBI Gene Expression Omnibus (GEO; GSE124819)34. This GEO project includes raw data in Fastq format, raw HTSeq counts28, and UCSC Genome Browser tracks in bigwig format for all samples23. This dataset is part of a larger study measuring the systemic transcriptional response to spinal cord injury, including dorsal root ganglion35 and soleus muscle, all of which are included as part of a GEO superseries (GSE129704)36.
Technical Validation
RNA metrics
Sequencing generated 26.5 to 46.8 million reads/sample with a mean of 37.1 million and standard deviation of 6.3 million. Table 1 displays the number of raw reads successfully aligned for each of the samples. The alignment rate for uniquely mapped and multi-mapped reads combined ranged from 85.97 to 98.87 percent with a mean of 97.85 across the 37 samples.
Quality assessment
Raw sequencing data was input to FastQC for quality assessment. All samples were deemed of high quality. In Fig. 5a, the Phred quality score per base is displayed for a representative sample from each experimental group. For all samples, the 25th percentile of quality scores is at or above a Phred score of 30, reflecting 99.9 percent accuracy in base calling. The gradual drop in quality at the end of the sequence is a common phenomenon with Illumina’s approach to sequencing by synthesis37.
Quality control analysis. Phred quality scores per base for one representative sample from each experimental group. (a) On the Y-axis a Phred score of 30 indicates 99.9% accuracy in base calling. Phred scores above 28 (green) are considered very good quality. Scores between 20 and 28 (orange) are considered reasonable quality. Scores below 20 (red) are considered poor quality. The yellow box represents the inter-quartile range (25–75%). The lower and upper whiskers represent the 10th and 90th percentiles respectively. On the right, 3D PCA plots for SCI + In-Cage Activity samples (b) SCI + Exercise samples minus TX SCI (c) and PCA focused solely on a comparison of injury severity (d).
Gene expression variation of biological replicates
PCA was performed to assess the within- and between-group variation of the samples. In Fig. 5b–d, three-dimensional PCA plots provide a view of sample points in 3D space. In Fig. 5b, the SCI + In-Cage Activity samples appear to have large variability within experimental groups (Variance: PC1 29%, PC2 13%, PC3 11%), reflecting some degree of individual differences across animals, one possible source being variability in the spontaneous activity of animals regardless of cage size. Importantly, however, the No SCI samples appear in a distinct region of the plot from the CONT SCI samples. A subset of samples from the CONT + Tiny and the CONT + Large groups are clearly distinct from each other with a few samples overlapping. The number of samples in each group, five each, would allow for removal of overlapping samples with a sufficient number of samples remaining, at least three, for comparison.
In Fig. 5c, PCA was performed to examine the separation between CONT SCI alone and CONT SCI followed by exercise (Variance: PC1 32%, PC2 13%, PC3 10%). The CONT + SWIM and CONT + SWW groups appear to overlap somewhat but are located in a distinct region of the graph from the CONT SCI and No SCI groups. Once again, sufficient samples are available in the SCI + Exercise groups to allow the removal of any overlapping samples between the SWW and SWIM groups. The CONT SCI samples cluster well and appear separate from the No SCI samples.
In Fig. 5d, PCA was performed to look at the separation between CONT SCI and TX SCI samples (Variance: PC1 28%, PC2 26%, PC3 9%). TX SCI samples vary widely within the group, but lie in a distinct location in the graph from the CONT SCI and the No SCI samples. In this case, there appears to be some overlap between the CONT SCI and the No SCI samples. However, by removing one sample from each group in the overlapping region, three samples remain distinctly separate in each group for comparison purposes.
Gene expression was examined to confirm that high level activity was found for genes relevant to liver function. In Fig. 3, the mean expression across CONT SCI samples is displayed for highly expressed genes associated with four well-documented liver pathologies. Mean expression for No SCI is included as a comparison. The genes associated with each category were obtained from topGO38. Figure 4 displays the UCSC Genome Browser expression tracks for the CONT SCI samples positioned at two genes known to be involved in lipid metabolism, Apoa1 (Fig. 4a) and Cyp1a2 (Fig. 4b). Expression appears consistent at all locations across the four CONT SCI animals.
Potential batch effects
The length of time between injury and tissue collection varied from 8.5 to 13.5 weeks in an attempt to balance the requirements of our research design with the well-being of the animals. The exercised animals required an initial period of time for introduction into the exercise facility followed by a full 10 weeks of exercise prior to measurement, resulting in tissue collection at 13.5 weeks post-injury. In contrast, the animals with a complete transection of the spinal cord required extensive care to ensure their well-being. Studies have shown that physiological measures plummet and stabilize by four weeks post-injury in the case of transection injury39. All batch effects were controlled since tissue was collected by the same individual using the same method and occurred well past the sub-acute to chronic stage transition which generally occurs 4 weeks post-injury40.
Code Availability
All analyses were performed using open sources software tools. Raw sequencing files were downloaded from Illumina BaseSpace using the Illumina Python Run Downloader41. Individual samples were initially divided across four lanes for sequencing, and these files were concatenated into one single end Fastq file using the UNIX cat command.
cat <FN1>.fastq <FN2>.fastq <FN3>.fastq <FN4>.fastq> <COND_REP>.fastq
The concatenated sequences were input to FastQC v.0.10.126 for quality control analysis using default parameters.
fastqc <COND_REP>.fastq –o <FASTQC_DIRECTORY>
Fastq files were input to Star 2.627 for alignment specifying BAM file format sorted by coordinate and requesting unmapped read files.
STAR–runMode alignReads–outSAMtype BAM SortedByCoordinate –outSAMstrandField intronMotif–outReadsUnmapped Fastx–readFilesIn <COND_REP>.fastq.gz –outFileNamePrefix <COND_REP>–runThreadN 16–genomeDir Rnor_6.0 –readFilesCommand zcat
Read counts for each sample were extracted using HTSeq 0.10.028. The reverse option was used to indicate strand orientation. Illumina’s TruSeq Stranded mRNA protocol was used to sequence the data and produces libraries where the first read is on the opposite strand to the RNA molecule.
htseq-count -f bam–stranded=reverse–mode=intersection-nonempty -r name <COND_REP>/Aligned.sortedByCoord.out.bam\Rattus_norvegicus.Rnor_6.0.93_PARSED.gtf> <COND_REP>/gene_counts_Reversed.htseq
The raw counts were normalized using DESeq2’s31,32 default procedure, relative log expression (RLE), using the estimateSizeFactors function. Detailed instructions can be found on the Bioconductor website for DESeq242.
References
Christopher & Dana Reeve Foundation. https://www.christopherreeve.org/ (2019).
Stein, D. M. et al. Risk factors for organ dysfunction and failure in patients with acute traumatic cervical spinal cord injury. Neurocrit. Care 13, 29–39, https://doi.org/10.1007/s12028-010-9359-9 (2010).
Nelson, M. D. et al. Metabolic syndrome in adolescents with spinal cord dysfunction. J. Spinal Cord Med. 30, S127–S139, https://doi.org/10.1080/10790268.2007.11754591 (2007).
Campbell, I. L. Cytokine-mediated inflammation, tumorigenesis, and disease-associated JAK/STAT/SOCS signaling circuits in the CNS. Brain Res. Rev. 48, 166–177, https://doi.org/10.1016/j.brainresrev.2004.12.006 (2005).
Gris, D., Hamilton, E. F. & Weaver, L. C. The systemic inflammatory response after spinal cord injury damages lungs and kidneys. Exp. Neurol. 211, 259–270, https://doi.org/10.1016/j.expneurol.2008.01.033 (2008).
Lenz, A., Franklin, G. A. & Cheadle, W. G. Systemic inflammation after trauma. Injury 38, 1336–1345, https://doi.org/10.1016/j.injury.2007.10.003 (2007).
Regel, G., Grotz, M., Weltner, T., Sturm, J. A. & Tscherne, H. Pattern of organ failure following severe trauma. World J. Surg. 20, 422–429, https://doi.org/10.1007/s002689900067 (1996).
Campbell, S. J. et al. Liver Kupffer cells control the magnitude of the inflammatory response in the injured brain and spinal cord. Neuropharmacology 55, 780–787, https://doi.org/10.1016/j.neuropharm.2008.06.074 (2008).
Hundt, H. et al. Assessment of hepatic inflammation after spinal cord injury using intravital microscopy. Injury 42, 691–696, https://doi.org/10.1016/j.injury.2010.12.013 (2011).
Fleming, J. C. et al. Remote inflammatory response in liver is dependent on the segmental level of spinal cord injury. J. Trauma Acute Care Surg. 72, 1194–1201, https://doi.org/10.1097/TA.0b013e31824d68bd (2012).
Barbonetti, A. et al. Low testosterone and non-alcoholic fatty liver disease: Evidence for their independent association in men with chronic spinal cord injury. J. Spinal Cord Med. 39, 443–449, https://doi.org/10.1179/2045772314Y.0000000288 (2016).
Sauerbeck, A. D. et al. Spinal cord injury causes chronic liver pathology in rats. J. Neurotrauma 32, 159–169, https://doi.org/10.1089/neu.2014.3497 (2015).
Jacobs, P. L., Mahoney, E. T., Cohn, K. A., Sheradsky, L. F. & Green, B. A. Oral creatine supplementation enhances upper extremity work capacity in persons with cervical-level spinal cord injury. Arch. Phys. Med. Rehabil. 83, 19–23, https://doi.org/10.1053/apmr.2002.26829 (2002).
Jacobs, P. L. & Nash, M. S. Exercise recommendations for individuals with spinal cord injury. Sports Med. 34, 727–751, https://doi.org/10.2165/00007256-200434110-00003 (2004).
Nash, M. S. Exercise as a health‐promoting activity following spinal cord injury. J. Neurol. Phys. Ther. 29(87–103), 106, https://doi.org/10.1097/01.NPT.0000282514.94093.c6 (2005).
Nash, M. S., van de Ven, I., van Elk, N. & Johnson, B. M. Effects of circuit resistance training on fitness attributes and upper-extremity pain in middle-aged men with paraplegia. Arch. Phys. Med. Rehabil. 88, 70–75, https://doi.org/10.1016/j.apmr.2006.10.003 (2007).
Caudle, K. L. et al. Hindlimb immobilization in a wheelchair alters functional recovery following contusive spinal cord injury in the adult rat. Neurorehab. Neural Repair 25, 729–739, https://doi.org/10.1177/1545968311407519 (2011).
Fouad, K., Metz, G. A., Merkler, D., Dietz, V. & Schwab, M. E. Treadmill training in incomplete spinal cord injured rats. Behav. Brain Res. 115, 107–113, https://doi.org/10.1016/S0166-4328(00)00244-8 (2000).
Kuerzi, J. et al. Task-specificity vs. ceiling effect: step-training in shallow water after spinal cord injury. Exp. Neurol. 224, 178–187, https://doi.org/10.1016/j.expneurol.2010.03.008 (2010).
Smith, R. R. et al. Effects of swimming on functional recovery after incomplete spinal cord injury in rats. J. Neurotrauma 23, 908–919, https://doi.org/10.1089/neu.2006.23.908 (2006).
Magnuson, D. S. et al. Swimming as a model of task-specific locomotor retraining after spinal cord injury in the rat. Neurorehabil. Neural Repair 23, 535–545, https://doi.org/10.1177/1545968308331147 (2009).
May, Z., Fouad, K., Shum-Siu, A. & Magnuson, D. S. K. Challenges of animal models in SCI research: Effects of pre-injury task-specific training in adult rats before lesion. Behav. Brain Res. 291, 26–35, https://doi.org/10.1016/j.bbr.2015.04.058 (2015).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006, https://doi.org/10.1101/gr.229102 (2002).
DeVeau, K. M. et al. A comparison of passive hindlimb cycling and active upper-limb exercise provides new insights into systolic dysfunction after spinal cord injury. Am. J. Physiol. Heart Circ. Physiol. 313, H861–H870, https://doi.org/10.1152/ajpheart.00046.2017 (2017).
Gruner, J. A. A monitored contusion model of spinal cord injury in the rat. J. Neurotrauma 9, 123–128, https://doi.org/10.1089/neu.1992.9.123 (1992).
Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, https://doi.org/10.1093/bioinformatics/bts635 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, https://doi.org/10.1093/bioinformatics/btu638 (2015).
Flicek, P. et al. Ensembl 2014. Nucleic Acids Res., gkt1196, https://doi.org/10.1093/nar/gkt1196 (2013).
The R Foundation. The R Project for Statistical Computing, https://www.r-project.org/ (2019).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106, https://doi.org/10.1186/gb-2010-11-10-r106 (2010).
Anders, S. et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8, 1765, https://doi.org/10.1038/nprot.2013.099 (2013).
Maza, E. In Papyro comparison of TMM (edgeR), RLE (DESeq2), and MRN normalization methods for a simple two-conditions-without-replicates RNA-Seq experimental design. Front. Genet. 7, 164, https://doi.org/10.3389/fgene.2016.00164 (2016).
Chariker, J. H. et al. Activity-induced changes in the liver transcriptome after chronic spinal cord injury. Gene Expression Omnibus, https://identifiers.org/geo:GSE124819 (2019).
Chariker, J. H. et al. Transcriptome of dorsal root ganglia caudal to a spinal cord injury with modulated behavioral activity. Sci. Data. 6, 88, https://doi.org/10.1038/s41597-019-0088-4 (2019).
Spinal cord injury. Gene Expression Omnibus, https://identifiers.org/geo:GSE129704 (2019).
Fuller, C. W. et al. The challenges of sequencing by synthesis. Nat. Biotechnol. 27, 1013, https://doi.org/10.1038/nbt.1585 (2009).
Alexa, A. & Rahnenfuhrer, J. Gene set enrichment analysis with topGO. Bioconductor Improv 27, http://bioconductor.riken.jp/packages/3.0/bioc/vignettes/topGO/inst/doc/topGO.pdf (2009).
Wang, F., Huang, S.-L., He, X.-J. & Li, X.-H. Determination of the ideal rat model for spinal cord injury by diffusion tensor imaging. Neuroreport 25, 1386, https://doi.org/10.1097/WNR.0000000000000278 (2014).
James, N. D. et al. Conduction failure following spinal cord injury: functional and anatomical changes from acute to chronic stages. J. Neurosci. 31, 18543–18555, https://doi.org/10.1523/JNEUROSCI.4306-11.2011 (2011).
Illumina. Illumina BaseSpace: Use the Python Run Downloader, https://help.basespace.illumina.com/articles/tutorials/using-the-python-run-downloader/ (2019).
Love, M.I., Anders, S. & Huber, W. Analyzing RNA-seq data with DESeq2, http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html (2019).
Acknowledgements
Support for J.H.C. and E.C.R. was provided by P20 GM103436 (Nigel Cooper, PI). Supported by the Leona and Harry Helmsley Charitable Trust (S.S.O., D.S.K.M., M.H., S.R.W., J.H.C., E.C.R.), Commonwealth of Kentucky Challenge for Excellence and P30 GM103507 (D.S.K.M., M.H., S.R.W.), and Norton Healthcare (SRW). We thank the University of Louisville Center for Genetics and Molecular Medicine (CGeMM) and the University of Louisville Genomics Core Facility, in particular Wolfgang Zacharias, Sabine Waigel, Ashley Wise, and Hong Gao for performing the high-throughput sequencing experiments. We thank Christine Yarberry, Johnny Morehouse, and Darlene Burke for surgical and behavioral analyses assistance.
Author information
Authors and Affiliations
Contributions
S.S.O., F.B., K.A.H., K.M.D. and D.S.K.M. planned all SCI experiments. S.S.O., F.B., K.A.H. and K.M.D. performed the SCI experiments. S.S.O., C.G., F.B. and K.A.H. collected and prepared tissue samples for RNA sequencing. J.H.C. and E.C.R. performed the bioinformatics analysis. J.H.C., S.S.O. and C.G. prepared the initial manuscript. All authors contributed to the manuscript editing process. Project funding was obtained as indicated above.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ISA-Tab metadata file
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Chariker, J.H., Ohri, S.S., Gomes, C. et al. Activity/exercise-induced changes in the liver transcriptome after chronic spinal cord injury. Sci Data 6, 88 (2019). https://doi.org/10.1038/s41597-019-0087-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-019-0087-5
This article is cited by
-
Knockdown of long non-coding RNA LEF1-AS1 attenuates apoptosis and inflammatory injury of microglia cells following spinal cord injury
Journal of Orthopaedic Surgery and Research (2021)
-
Transcriptome of dorsal root ganglia caudal to a spinal cord injury with modulated behavioral activity
Scientific Data (2019)