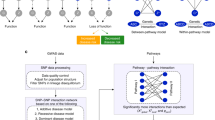
Abstract
Genetic interactions have the potential to modulate phenotypes, including human disease. In principle, genome-wide association studies (GWAS) provide a platform for detecting genetic interactions; however, traditional methods for identifying them, which tend to focus on testing individual variant pairs, lack statistical power. In this protocol, we describe a novel computational approach, called Bridging Gene sets with Epistasis (BridGE), for discovering genetic interactions between biological pathways from GWAS data. We present a Python-based implementation of BridGE along with instructions for its application to a typical human GWAS cohort. The major stages include initial data processing and quality control, construction of a variant-level genetic interaction network, measurement of pathway-level genetic interactions, evaluation of statistical significance using sample permutations and generation of results in a standardized output format. The BridGE software pipeline includes options for running the analysis on multiple cores and multiple nodes for users who have access to computing clusters or a cloud computing environment. In a cluster computing environment with 10 nodes and 100 GB of memory per node, the method can be run in less than 24 h for typical human GWAS cohorts. Using BridGE requires knowledge of running Python programs and basic shell script programming experience.
Key points
-
This protocol describes a method for discovering interactions between biological pathways in genome-wide association study data by evaluating variant-level interactions connecting between and within biological pathways.
-
The technique differs from approaches that perform interaction tests for every pair of variants with a phenotype of interest as it specifically assesses the impact of combinations of interacting loci at the pathway level, which affords Bridging Gene sets with Epistasis (BridGE) greater statistical power for identifying genetic interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The datasets used in this protocol included (1) a sample 1000 Genomes Project GWAS dataset with simulated binary phenotypes, which is available at the Zenodo repository (https://doi.org/10.5281/zenodo.8067407); (2) a copy of 1000 Genomes Project data, which is available at the Zenodo repository (https://doi.org/10.5281/zenodo.8067407); and (3) a Parkinson’s disease GWAS dataset from the IPDGC (dbGaP study accession: phs000918.v1.p1), which is available at the dbGaP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000918.v1.p1).
Code availability
BridGE-Python can be obtained from the GitHub repository (https://github.com/csbio/BridGE-Python). It can be freely used for educational and research purposes by nonprofit institutions and US government agencies. A license for commercial use of this software is available from the University of Minnesota’s Office for Technology Commercialization.
References
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Eichler, E. E. et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11, 446–450 (2010).
Maher, B. Personal genomes: the case of the missing heritability. Nature 456, 18–21 (2008).
Phillips, P. C. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9, 855–867 (2008).
Cordell, H. J. Detecting gene–gene interactions that underlie human diseases. Nat. Rev. Genet. 10, 392–404 (2009).
Mackay, T. F. & Moore, J. H. Why epistasis is important for tackling complex human disease genetics. Genome Med. 6, 42 (2014).
Zuk, O., Hechter, E., Sunyaev, S. R. & Lander, E. S. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl Acad. Sci. USA 109, 1193–1198 (2012).
Hu, X. et al. SHEsisEpi, a GPU-enhanced genome-wide SNP–SNP interaction scanning algorithm, efficiently reveals the risk genetic epistasis in bipolar disorder. Cell Res. 20, 854–857 (2010).
Schüpbach, T., Xenarios, I., Bergmann, S. & Kapur, K. FastEpistasis: a high performance computing solution for quantitative trait epistasis. Bioinformatics 26, 1468–1469 (2010).
Wan, X. et al. BOOST: a fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am. J. Hum. Genet. 87, 325–340 (2010).
Yung, L. S., Yang, C., Wan, X. & Yu, W. GBOOST: a GPU-based tool for detecting gene–gene interactions in genome-wide case control studies. Bioinformatics 27, 1309–1310 (2011).
Goudey, B. et al. GWIS-model-free, fast and exhaustive search for epistatic interactions in case-control GWAS. BMC Genom. 14, 1–18 (2013).
Wang, X. et al. ELSSI: parallel SNP–SNP interactions detection by ensemble multi-type detectors. Brief. Bioinform. 23, bbac213 (2022).
Chatelain, C., Durand, G., Thuillier, V. & Augé, F. Performance of epistasis detection methods in semi-simulated GWAS. BMC Bioinform. 19, 1–17 (2018).
Costanzo, M. et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420 (2016).
Kuzmin, E. et al. Systematic analysis of complex genetic interactions. Science 360, eaao1729 (2018).
Wang, W. et al. Pathway-based discovery of genetic interactions in breast cancer. PLoS Genet. 13, e1006973 (2017).
Fang, G. et al. Discovering genetic interactions bridging pathways in genome-wide association studies. Nat. Commun. 10, 4274 (2019).
Ueki, M. & Cordell, H. J. Improved statistics for genome-wide interaction analysis. PLoS Genet. 8, e1002625 (2012).
Sollis, E. et al. The NHGRI-EBI GWAS catalog: knowledgebase and deposition resource. Nucleic Acids Res. 51, D977–D985 (2023).
Hallacli, E. et al. The Parkinson’s disease protein alpha-synuclein is a modulator of processing bodies and mRNA stability. Cell 185, 2035–2056.e33 (2022).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Wang, K., Li, M. & Bucan, M. Pathway-based approaches for analysis of genome-wide association studies. Am. J. Hum. Genet. 81, 1278–1283 (2007).
Kim, N. C. et al. Gene ontology analysis of pairwise genetic associations in two genome-wide studies of sporadic ALS. BioData Min. 5, 9 (2012).
Pandey, A. et al. Epistasis network centrality analysis yields pathway replication across two GWAS cohorts for bipolar disorder. Transl. Psychiatry 2, e154–e154 (2012).
Ma, L. et al. Knowledge-driven analysis identifies a gene–gene interaction affecting high-density lipoprotein cholesterol levels in multi-ethnic populations. PLoS Genet. 8, e1002714 (2012).
Ma, L., Clark, A. G. & Keinan, A. Gene-based testing of interactions in association studies of quantitative traits. PLoS Genet. 9, e1003321 (2013).
Sun, X. et al. Analysis pipeline for the epistasis search–statistical versus biological filtering. Front. Genet. 5, 106 (2014).
Brossard, M. et al. Integrated pathway and epistasis analysis reveals interactive effect of genetic variants at TERF1 and AFAP1L2 loci on melanoma risk. Int. J. Cancer 137, 1901–1909 (2015).
Mitra, I. et al. Reverse pathway genetic approach identifies epistasis in autism spectrum disorders. PLoS Genet. 13, e1006516 (2017).
Chen, L. S. et al. Insights into colon cancer etiology via a regularized approach to gene set analysis of GWAS data. Am. J. Hum. Genet. 86, 860–871 (2010).
Zhao, J., Gupta, S., Seielstad, M., Liu, J. & Thalamuthu, A. Pathway-based analysis using reduced gene subsets in genome-wide association studies. BMC Bioinform. 12, 1–14 (2011).
Huang, A., Martin, E. R., Vance, J. M. & Cai, X. Detecting genetic interactions in pathway‐based genome‐wide association studies. Genet. Epidemiol. 38, 300–309 (2014).
Ritchie, M. D. Large-scale analysis of genetic and clinical patient data. Annu. Rev. Biomed. Data Sci. 1, 263–274 (2018).
Silberstein, M., Nesbit, N., Cai, J. & Lee, P. H. Pathway analysis for genome-wide genetic variation data: analytic principles, latest developments, and new opportunities. J. Genet. Genom. 48, 173–183 (2021).
Cui, T. et al. Gene–gene interaction detection with deep learning. Commun. Biol. 5, 1238 (2022).
Liu, L. et al. Using machine learning to identify gene interaction networks associated with breast cancer. BMC Cancer 22, 1070 (2022).
Consortium, G. P. A global reference for human genetic variation. Nature 526, 68 (2015).
Consortium, I. P. D. G. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet 377, 641–649 (2011).
Lewontin, R. C. & Kojima, K.-I. The evolutionary dynamics of complex polymorphisms. Evolution 14, 458–472 (1960).
Acknowledgements
This work was partially supported by a grant from the Alzheimer’s Association, Alzheimer’s Research UK, The Michael J. Fox Foundation for Parkinson’s Research and the Weston Brain Institute (BAND-19-615151), and grants from the NIH (R21CA235352, R01HG005084 and R01HG005853). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders. This study makes use of genome-wide association datasets provided by dbGaP (study accession numbers: phs000089.v3.p2, phs000126.v2.p1, phs000918.v1.p1). We acknowledge the contributing investigators who submitted data from their original study to dbGaP, the primary funding organization that supported the contributing investigators and the NIH data repository. Computing resources and data storage services were partially provided by the Minnesota Supercomputing Institute and the University of Minnesota’s Office of Information Technology, respectively.
Author information
Authors and Affiliations
Contributions
M.H., W.W. and C.L.M. created the software design based on the original BridGE method. M.H., W.W. and M.A. developed the software. M.H., M.F. and W.W. tested the protocol and provided feedback on improvements. M.H. and W.W. wrote an initial draft of the protocol manuscript, which was then reviewed and revised by all co-authors. W.W. and C.L.M. supervised the software development process, manuscript writing and testing of the protocol, and secured funding to support the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Marylyn Ritchie and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Wang, W. et al. PLoS Genet. 13, e1006973 (2017): https://doi.org/10.1371/journal.pgen.1006973
Fang, G. et al. Nat. Commun. 10, 4274 (2019): https://doi.org/10.1038/s41467-019-12131-7
Supplementary information
Supplementary Data 1
The output of the original BridGE and BridGE 2.0 are compared for the two Parkinson’s disease GWAS cohorts, which were also analyzed in the original BridGE paper. The Excel files include summary tables and lists of BPM, WPM and PATH discovered at the FDR cutoff shown in the summary tables.
Supplementary Data 2
BridGE output files from the simulated 1000 Genomes GWAS data based on the combined disease model. The Excel files include BridGE results and interaction lists for BPMs and WPMs. The PDF file provides the visualization of pathway–pathway interactions.
Supplementary Data 3
A complete set of BridGE output files from the Parkinson’s disease IPDGC cohort (dbGaP study accession: phs000918.v1.p1). The Excel files include information about significant pathway-level interactions (FDR <0.25) discovered under the corresponding disease model (RR, DD, RD and combined) and pairwise SNP interaction lists and associated statistics (DD disease model only), one for BPM and one for WPM. The PDF file provides a network visualization.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hajiaghabozorgi, M., Fischbach, M., Albrecht, M. et al. BridGE: a pathway-based analysis tool for detecting genetic interactions from GWAS. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-00954-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-00954-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.