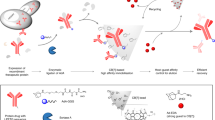
Abstract
Monoclonal antibodies (mAbs) are commonly used biologic drugs for the treatment of diseases such as rheumatoid arthritis, multiple sclerosis, COVID-19 and various cancers. They are produced in Chinese hamster ovary cell lines and are purified via a number of complex and expensive chromatography-based steps, operated in batch mode, that rely heavily on protein A resin. The major drawback of conventional procedures is the high cost of the adsorption media and the extensive use of chemicals for the regeneration of the chromatographic columns, with an environmental cost. We have shown that conventional protein A chromatography can be replaced with a single crystallization step and gram-scale production can be achieved in continuous flow using the template-assisted membrane crystallization process. The templates are embedded in a membrane (e.g., porous polyvinylidene fluoride with a layer of polymerized polyvinyl alcohol) and serve as nucleants for crystallization. mAbs are flexible proteins that are difficult to crystallize, so it can be challenging to determine the optimal conditions for crystallization. The objective of this protocol is to establish a systematic and flexible approach for the design of a robust, economic and sustainable mAb purification platform to replace at least the protein A affinity stage in traditional chromatography-based purification platforms. The procedure provides details on how to establish the optimal parameters for separation (crystallization conditions, choice of templates, choice of membrane) and advice on analytical and characterization methods.
Key points
-
Monoclonal antibodies used in clinical applications need to be produced and purified at a gram scale. Purification by crystallization is less expensive and more environmentally sustainable than the standard chromatography approach. However, finding appropriate crystallization conditions is challenging.
-
In template-assisted crystallization, the template is a heteronucleant specifically designed to induce crystallization. In this approach, the template is attached to a membrane that is used, at scale, for osmotic membrane-assisted crystallization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout













Similar content being viewed by others
Data availability
Data supporting this publication are reported in ZENODO under reference codes https://doi.org/10.5281/zenodo.3856655, https://doi.org/10.5281/zenodo.3874571, https://doi.org/10.5281/zenodo.3245362, https://doi.org/10.5281/zenodo.2605199 and https://doi.org/10.5281/zenodo.1322662, and in the database SASBDB at the entry SASDMX3.
References
Hughson, M. D., Cruz, T. A., Carvalho, R. J. & Castilho, L. R. Development of a 3-step straight-through purification strategy combining membrane adsorbers and resins. Biotechnol. Prog. 33, 931–940 (2017).
Trnovec, H., Doles, T., Hribar, G., Furlan, N. & Podgornik, A. Characterization of membrane adsorbers used for impurity removal during the continuous purification of monoclonal antibodies. J. Chromatogr. A 1609, 460518 (2020).
Shukla, A. A., Wolfe, L. S., Mostafa, S. S. & Norman, C. Evolving trends in mAb production processes. Bioeng. Transl. Med. 2, 58–69 (2017).
Liu, H. F., Ma, J., Winter, C. & Bayer, R. Recovery and purification process development for monoclonal antibody production. mAbs 2, 480–499 (2010).
dos Santos, R., Carvalho, A. L. & Roque, A. C. A. Renaissance of protein crystallization and precipitation in biopharmaceuticals purification. Biotechnol. Adv. 35, 41–50 (2017).
Curtis, S., Lee, K., Blank, G. S., Brorson, K. & Xu, Y. Generic/matrix evaluation of SV40 clearance by anion exchange chromatography in flow-through mode. Biotechnol. Bioeng. 84, 179–186 (2003).
Norling, L. et al. Impact of multiple re-use of anion-exchange chromatography media on virus removal. J. Chromatogr. A 1069, 79–89 (2005).
Liu, H. F. et al. Exploration of overloaded cation exchange chromatography for monoclonal antibody purification. J. Chromatogr. A 1218, 6943–6952 (2011).
Porath, J. Salt-promoted adsorption: recent developments. J. Chromatogr. B 376, 331–341 (1986).
Roper, D. K. & Lightfoot, E. N. Separation of biomolecules using adsorptive membranes. J. Chromatogr. A 702, 3–26 (1995).
Knudsen, H. L., Fahrner, R. L., Xu, Y., Norling, L. A. & Blank, G. S. Membrane ion-exchange chromatography for process-scale antibody purification. J. Chromatogr. A 907, 145–154 (2001).
Lewis, U. J., Williams, D. E. & Brink, N. G. Pancreatic elastase: purification, properties, and function. J. Biol. Chem. 222, 705–720 (1956).
Srivastava, O. P. & Aronson, A. I. Isolation and characterization of a unique protease from sporulating cells of Bacillus subtilis. Arch. Microbiol. 129, 227–232 (1981).
Judge, R. A., Johns, M. R. & White, E. T. Protein purification by bulk crystallization: the recovery of ovalbumin. Biotechnol. Bioeng. 48, 316–323 (1995).
Jacobsen, C., Garside, J. & Hoare, M. Nucleation and growth of microbial lipase crystals from clarified concentrated fermentation broths. Biotechnol. Bioeng. 57, 666–675 (1998).
Peters, J., Minuth, T. & Schröder, W. Implementation of a crystallization step into the purification process of a recombinant protein. Protein Expr. Purif. 39, 43–53 (2005).
Shah, U. V., Amberg, C., Diao, Y., Yang, Z. & Heng, J. Y. Heterogeneous nucleants for crystallogenesis and bioseparation. Curr. Opin. Chem. Eng. 8, 69–75 (2015).
Zang, Y., Kammerer, B., Eisenkolb, M., Lohr, K. & Kiefer, H. Towards protein crystallization as a process step in downstream processing of therapeutic antibodies: screening and optimization at microbatch scale. PLoS One 6, e25282 (2011).
Smejkal, B. et al. Fast and scalable purification of a therapeutic full-length antibody based on process crystallization. Biotechnol. Bioeng. 110, 2452–2461 (2013).
Sommer, R. et al. Combined polyethylene glycol and CaCl2 precipitation for the capture and purification of recombinant antibodies. Process Biochem. 49, 2001–2009 (2014).
Oelmeier, S. A., Ladd-Effio, C. & Hubbuch, J. Alternative separation steps for monoclonal antibody purification: combination of centrifugal partitioning chromatography and precipitation. J. Chromatogr. A 1319, 118–126 (2013).
Hammerschmidt, N., Hobiger, S. & Jungbauer, A. Continuous polyethylene glycol precipitation of recombinant antibodies: sequential precipitation and resolubilization. Process Biochem. 51, 325–332 (2016).
Sommer, R. et al. A. Capture and intermediate purification of recombinant antibodies with combined precipitation methods. Biochem. Eng. J. 93, 200–211 (2015).
Sim, S.-L. et al. Branched polyethylene glycol for protein precipitation. Biotechnol. Bioeng. 109, 736–746 (2012).
Yang, H. et al. Optimization of vapor diffusion conditions for anti-CD20 crystallization and scale-up to Meso Batch. Crystals 9, 230 (2019).
McPherson, A. & Shlichta, P. Heterogeneous and epitxial nucleation of protein crystal on mineral surfaces. Science 239, 385–387 (1988).
Hodzhaoglu, F. K., Kurniawan, F., Mirsky, V. & Nanev, C. Gold nanoparticles induce protein crystallization. Cryst. Res. Technol. 43, 588–593 (2008).
Dobrev, D., Baur, D. & Neumann, R. Crystallization of lysozyme in pores of etched heavy-ion tracks. Appl. Phys. A 80, 451–456 (2005).
Shah, U., Williams, D. & Heng, J. Y. Y. Selective crystallization of proteins using engineered nanonucleants. Cryst. Growth Des. 12, 1362–1369 (2012).
Chen, W. et al. High protein-loading silica template for heterogeneous protein crystallization. Cryst. Growth Des. 20, 866–873 (2020).
Bracewell, D. G., Pathak, M., Ma, G. & Rathore, A. S. Re-use of protein A resin: fouling and economics. BioPharm. Int. 28, 28–33 (2015).
Curcio, E., Di Profio, G. & Drioli, E. Membrane crystallization of macromolecular solutions. Desalination 145, 173 (2002).
Curcio, E., Di Profio, G. & Drioli, E. Recovery of fumaric acid by membrane crystallization in the production of l-malic acid. Sep. Pur. Technol. 33, 63 (2003).
Curcio, E., Di Profio, G. & Drioli, E. A new membrane-based crystallization technique: tests on lysozyme. J. Cryst. Growth 247, 166–176 (2003).
Curcio, E., Fontananova, E., Di Profio, G. & Drioli, E. Influence of the structural properties of poly(vinylidene fluoride) membranes on the heterogeneous nucleation rate of protein crystals. J. Phys. Chem. B 110, 12438 (2006).
Curcio, E. et al. Membrane distillation operated at high seawater concentration factors: role of the membrane on CaCO3 scaling in presence of humic acid. J. Membr. Sci. 346, 263 (2010).
Curcio, E. et al. Membrane crystallization of lysozyme under forced solution flow. J. Membr. Sci. 257, 134 (2005).
Di Profio, G. et al. Fine dosage of antisolvent in the crystallization of l-histidine: effect on polymorphism. Cryst. Growth Des. 10, 449 (2010).
Di Profio, G., Curcio, E., Cassetta, A., Lamba, D. & Drioli, E. Membrane crystallization of lysozyme: kinetic aspects. J. Cryst. Growth 257, 359 (2003).
Di Profio, G., Curcio, E. & Drioli, E. Trypsin crystallization by membrane-based techniques. J. Struct. Biol. 150, 41 (2005).
Di Profio, G., Curcio, E. & Drioli, E. Supersaturation control and heterogeneous nucleation in membrane crystallizers: facts and perspectives. Ind. Eng. Chem. Res. 49, 11878–11889 (2010).
Di Profio, G., Curcio, E., Ferraro, S., Stabile, C. & Drioli, E. Effect of supersaturation control and heterogeneous nucleation on porous membrane surfaces in the crystallization of l-glutamic acid polymorphs. Cryst. Growth Des. 9, 2179 (2009).
Di Profio, G. et al. Preparation of enzyme crystals with tunable morphology in membrane crystallizers. Ind. Eng. Chem. Res. 44, 10005 (2005).
Di Profio, G., Stabile, C., Caridi, A., Curcio, E. & Drioli, E. Antisolvent membrane crystallization of pharmaceutical compounds. J. Pharm. Sci. 98, 4902 (2009).
Di Profio, G., Tucci, S., Curcio, E. & Drioli, E. Selective glycine polymorph crystallization by using microporous membranes. Cryst. Growth Des. 7, 526 (2007).
Di Profio, G., Tucci, S., Curcio, E. & Drioli, E. Controlling polymorphism with membrane-based crystallizers: application to form I and II of paracetamol. Chem. Mater. 19, 2386 (2007).
Ding, Z., Liu, L., El-Bourawi, M. S. & Ma, R. Analysis of a solar-powered membrane distillation system. Desalination 172, 27 (2005).
Drioli, E., Curcio, E., Criscuoli, A. & Di Profio, G. Integrated system for recovery of CaCO3, NaCl and MgSO4·7H2O from nanofiltration retentate. J. Membr. Sci. 239, 27 (2004).
Drioli, E., Curcio, E. & Di Profio, G. State of the art and recent progresses in membrane contactors. Chem. Eng. Res. Des. 83, 223 (2005).
Drioli, E., Curcio, E., Di Profio, G., Macedonio, F. & Criscuoli, A. Integrating membrane contactors technology and pressure-driven membrane operations for seawater desalination: energy, exergy and costs analysis. Chem. Eng. Res. Des. 84, 209 (2006).
Di Profio, G. et al. Tailored hydrogel membranes for efficient protein crystallization. Adv. Funct. Mater. 24, 1582–1590 (2014).
Drioli, E., Di Profio, G., Curcio, E. Membrane-Assisted Crystallization Technology, Advances in Chemical and Process Engineering 2 (Imperial College Press, 2015).
Polino, M., Portugal, C. A. M., Di Profio, G., Coelhoso, I. M. & Crespo, J. G. Protein crystallization by membrane-assisted technology. Cryst. Growth Des. 19, 4871–4883 (2019).
Roque, A. C. A. et al. Anything but conventional chromatography approaches in bioseparation. Biotechnol. J. 15, 1900274 (2020).
Slabinski, L. et al. A web server for prediction of protein crystallizability. Bioinformatics 23, 3403–3405 (2008).
Kurgan, L. et al. CRYSTALP2: sequence-based protein crystallization propensity prediction. BMC Struct. Biol. 9, 50 (2009).
Goldschmidt, L., Derewenda, D. C. Z. & Eisenberg, D. Toward rational protein crystallization: a Web server for the design of crystallizable protein variants. Protein Sci. 16, 1569–1576 (2007).
Hammadi, Z. et al. Localizing and inducing primary nucleation. Faraday Discuss. 179, 489–501 (2015).
Pantuso, E. et al. On the aggregation and nucleation mechanism of the monoclonal antibody anti-CD20 near liquid-liquid phase separation (LLPS). Sci. Rep. 10, 8902 (2020).
Chan, H. Y. & Lubchenko, V. A mechanism for reversible mesoscopic aggregation in liquid solutions. Nat. Commun. 10, 2381 (2019).
Shah, U. V. et al. Crystallization via novel 3D nanotemplates as a tool for protein purification and bio-separation. J. Cryst. Growth 469, 42–47 (2017).
Muschol, M. & Rosenberger, F. Liquid–liquid phase separation in supersaturated lysozyme solutions and associated precipitate formation/crystallization. J. Chem. Phys. 107, 1953–1962 (1997).
Chari, R., Jerath, K., Badkar, A. V. & Kalonia, D. S. Long- and short-range electrostatic interactions affect the rheology of highly concentrated antibody solutions. Pharm. Res. 26, 2607 (2009).
Mahler, H.-C., Friess, W., Grauschopf, U. & Kiese, S. Protein aggregation: pathways, induction factors and analysis. J. Pharm. Sci. 98, 2909 (2009).
Trilisky, E., Gillespie, R., Osslund, T. D. & Vunnum, S. Crystallization and liquid-liquid phase separation of monoclonal antibodies and Fc-fusion proteins: screening results. Biotechnol. Prog. 27, 1054–1067 (2011).
Lewus, R. A., Darcy, P. A., Lenhoff, A. M. & Sandler, S. I. Interactions and phase behavior of a monoclonal antibody. Biotechnol. Prog. 27, 280–289 (2011).
Dumetz, A. C., Lewus, R. A., Lenhoff, A. M. & Kaler, E. W. Effects of ammonium sulfate and sodium chloride concentration on PEG/protein liquid-liquid phase separation. Langmuir 24, 10345–10351 (2008).
Roberts, C. J., Das, T. K. & Sahin, E. Predicting solution aggregation rates for therapeutic proteins: approaches and challenges. Int. J. Pharm. 418, 318 (2011).
Velev, O. D., Kaler, E. W. & Lenhoff, A. M. Protein interactions in solution characterized by light and neutron scattering: comparison of lysozyme and chymotrypsinogen. Biophys. J. 75, 2682 (1998).
Neal, B. L., Asthagiri, D. & Lenhoff, A. M. Molecular origins of osmotic second virial coefficients of proteins. Biophys. J. 75, 2469 (1998).
George, A. & Wilson, W. W. Predicting protein crystallization from a dilute solution property. Acta Crystallogr. D. 50, 361–365 (1994).
Haas, C. & Drenth, J. The protein–water phase diagram and the growth of protein crystals from aqueous solution. J. Phys. Chem. B 102, 4226 (1998).
Hassan, P. A., Rana, S. & Verma, G. Making sense of Brownian motion: colloid characterization by dynamic light scattering. Langmuir 31, 3–12 (2015).
Zhang, J. & Liu, X. Y. Effect of protein–protein interactions on protein aggregation kinetics. J. Chem. Phys. 119, 10972–10976 (2003).
Harding, S. E. & Johnson, P. The concentration-dependence of macromolecular parameters. Biochem. J. 231, 543–547 (1985).
Wilson, W. W. Monitoring crystallization experiments using dynamic light scattering: assaying and monitoring protein crystallization in solution. Methods 1, 110–117 (1990).
Wu, G. et al. Elucidating the weak protein–protein interaction mechanisms behind the liquid–liquid phase separation of a mAb solution by different types of additives. Eur. J. Pharm. Biopharm. 120, 1–8 (2017).
Veesler, S., Revalor, E., Bottini, O. & Hoff, C. Crystallization in the presence of a liquid–liquid phase separation. Org. Process Res. Dev. 10, 841–845 (2006).
Duffy, D. et al. In situ monitoring, control and optimization of a liquid–liquid phase separation crystallization. Chem. Eng. Sci. 77, 112–121 (2012).
Chen, W., Karde, V., Cheng, T. N., Ramli, S. S. & Heng, J. Y. Surface hydrophobicity: effect of alkyl chain length and network homogeneity. Front. Chem. Sci. Eng. 15, 1–9 (2020).
Chayen, N. E., Saridakis, E., El-Bahar, R. & Nemirovsky, Y. Porous silicon: an effective nucleation-inducing material for protein crystallization. J. Mol. Biol. 312, 591–595 (2001).
Chayen, N. E., Saridakis, E. & Sear, R. P. Experiment and theory for heterogeneous nucleation of protein crystals in a porous medium. Proc. Natl Acad. Sci. USA 103, 597–601 (2006).
Page, A. J. & Sear, R. P. Heterogeneous nucleation in and out of pores. Phys. Rev. Lett. 97, 065701 (2006).
Artusio, F. & Pisano, R. Surface-induced crystallization of pharmaceuticals and biopharmaceuticals: a review. Int. J. Pharm. 547, 190–208 (2018).
Nanev, C. N., Saridakis, E. & Chayen, N. E. Protein crystal nucleation in pores. Sci. Rep. 7, 35821 (2017).
Lam, J. & Lutsko, J. F. Solute particle near a nanopore: influence of size and surface properties on the solvent-mediated forces. Nanoscale 9, 17099–17108 (2017).
Zhang, S.-Q. & Cheung, M. S. Manipulating biopolymer dynamics by anisotropic nanoconfinement. Nano Lett. 7, 3438–3442 (2007).
van Meel, J. A., Sear, R. P. & Frenkel, D. Design principles for broad-spectrum protein-crystal nucleants with nanoscale pits. Phys. Rev. Lett. 105, 205501 (2010).
Galkin, O. & Vekilov, P. G. Nucleation of protein crystals: critical nuclei, phase behavior, and control pathways. J. Cryst. Growth 232, 63–76 (2001).
Leng, J. & Salmon, J.-B. Microfluidic crystallization. Lab Chip 9, 24–34 (2009).
Li, L. & Ismagilov, R. F. Protein crystallization using microfluidic technologies based on valves, droplets, and SlipChip. Annu. Rev. Biophys. 39, 139–158 (2010).
Candoni, N., Grossier, R., Lagaize, M. & Veesler, S. Advances in the use of microfluidics to study crystallization fundamentals. Ann. Rev. Chem. Biomol. Eng. 10, 59–83 (2019).
Gerard, C. J. J. et al. Template-assisted crystallization behavior in stirred solutions of the monoclonal antibody anti-CD20: probability distributions of induction times. Cryst. Growth Des. 22, 3637–3645 (2022).
McPherson, A. in Protein Crystallography (eds. Wlodawer, A., Dauter, Z. & Jaskolski, M.) 17–50 (Springer, 2017).
Strathmann, H., Giorno, L. & Drioli, E. in Comprehensive Membrane (eds. Drioli, E., Giorno, L.) 91−111 (Elsevier, 2010).
Figoli, A. et al. in Membrane Fabrication (eds. Hilal, N., Ismail, A. F. & Wright, C.) (CRC Press, 2015).
Meringolo, C. et al. Tailoring PVDF membranes surface topography and hydrophobicity by a sustainable two-steps phase separation process. ACS Sustain. Chem. Eng. 6, 10069–10077 (2018).
Salehi, S. M. et al. Hydrogel composite membranes incorporating iron oxide nanoparticles as topographical designers for controlled heteronucleation of proteins. Cryst. Growth Des. 18, 3317–3327 (2018).
Meringolo, C. et al. Exploiting fluoropolymers immiscibility to tune surface properties and mass transfer in blend membranes for membrane contactor applications. ACS Appl. Polym. Mater. 1, 326–334 (2019).
Curcio, E., Curcio, V., Di Profio, G., Fontananova, E. & Drioli, E. Energetics of protein nucleation on rough polymeric surfaces. J. Phys. Chem. B 114, 13650–13655 (2010).
Cassie, B. D. B. & Baxter, S. Wettability of porous surfaces. Trans. Faraday Soc. 40, 546–551 (1944).
Curcio, E., Di Profio, G. & Drioli, E. in Comprehensive Membrane Science and Engineering Vol. 4, Ch. 4.01, 1−20 (Elsevier, 2010).
Alkhudhiri, A., Darwish, N. & Hilal, H. Membrane distillation: a comprehensive review. Desalination 287, 2–18 (2012).
Liu, Y.-X., Wang, X.-J., Lu, J. & Ching, C. B. Influence of the roughness, topography, and physicochemical properties of chemically modified surfaces on the heterogeneous nucleation of protein crystals. J. Phys. Chem. B 111, 13971–13978 (2007).
Drioli, E., Criscuoli, A. & Curcio, E. Membrane Contactors: Fundamentals Applications and Potentialities (Elsevier, 2011).
Vitola, G., Mazzei, R., Fontananova, E. & Giorno, L. PVDF membrane biofunctionalization by chemical grafting. J. Membr. Sci. 476, 483–489 (2015).
Liu, F., Hashim, N. A., Liu, Y., Abed, M. R. M. & Li, K. Progress in the production and modification of PVDF membranes. J. Membr. Sci. 375, 1–27 (2011).
Patterson, A. The Scherrer formula for X-ray particle size determination. Phys. Rev. 56, 978–982 (1939).
The CMC Biotech Working Group. A-Mab: a case study in bioprocess development. https://qbdworks.com/storage/2014/06/A-MabCaseStudyVersion.pdf (2009).
Lacher, N. A. et al. Development, validation, and implementation of capillary gel electrophoresis as a replacement for SDS-PAGE for purity analysis of IgG2 mAbs. J. Sep. Sci. 33, 218–227 (2010).
Yao, H., Wynendaele, E., Xu, X., Kosgei, A. & De Spiegeleer, B. Circular dichroism in functional quality evaluation of medicines. J. Pharm. Biomed. Anal. 147, 50–64 (2018).
Tscheliessnig, A. L., Konrath, J., Bates, R. & Jungbauer, A. Host cell protein analysis in therapeutic protein bioprocessing–methods and applications. Biotechnol. J. 8, 655–670 (2013).
Ruppach, H. Log10 reduction factors in viral clearance studies. BioProcess. J. 12, 24–30 (2014).
Orchard, J. D. et al. Using a noninfectious MVM surrogate for assessing viral clearance during downstream process development. Biotechnol. Prog. 36, e2921 (2019).
Dreckmann, T., Boeuf, J., Ludwig, I.-S., Lümkemann, J. & Huwyler, J. Low volume aseptic filling: Impact of pump systems on shear stress. Eur. J. Pharm. Biopharm. 147, 10–18 (2020).
Sparks, T. C. in Filters and Filtration Handbook 5th edn. (eds. Sparks, T. & Chase, G.) Section 5, 297–359 (Butterworth–Heinemann, 2016).
Dao, L., Nguyen, H. & Mai, H. A fiber optic turbidity system for in-situ monitoring protein aggregation, nucleation and crystallization. Acta Astronautica 47, 399–409 (2000).
Huffman, S., Soni, K. & Ferraiolo, J. UV-Vis based determination of protein concentration: Validating and implementing slope measurements using variable pathlength technology. Bioprocess. Int. 12, 66–72 (2014).
AMECRYS. Deliverable D2.2 HEL4 domain fragment & anti-CD20 mAb process specification report. https://ec.europa.eu/research/participants/documents/downloadPublic?documentIds=080166e5b99e2bac&appId=PPGMS (2018).
Grottshalk, U. Process Scale Purification of Antibodies 2nd edn. (John Wiley, 2017).
Godavarti, R., Tugcu, N., Kumar, V. & Rathore, A. Evolution of the monoclonal antibody purification platform. BioPharm. Int. 26, 32–37 (2013).
Cytiva. Affinity Chromatography Vol. 1 https://cdn.cytivalifesciences.com/api/public/content/digi-11660-pdf (2021).
Smith, M. R. Rituximab (monoclonal anti-CD20 antibody): mechanisms of action and resistance. Oncogene 22, 7359–7368 (2003).
Pescovitz, M. D. Rituximab, an anti-CD20 monoclonal antibody: history and mechanism of action. Am. J. Transplant. 6, 859–866 (2006).
de Araujo Sampaio, D. et al. Aqueous two-phase (polyethylene glycol+sodium sulfate) system for caffeine extraction: equilibrium diagrams and partitioning study. J. Chem. Thermodynam. 98, 86–94 (2016).
Chayen, N. E., Shaw Stewart, P. D., Maeder, D. L. & Blow, D. M. An automated system for micro-batch protein crystallization and screening. J. Appl. Crystallogr. 23, 297–302 (1990).
Carter, C. W. & Carter, C. W. Protein crystallization using incomplete factorial-experiments. J. Biol. Chem. 254, 2219–2223 (1979).
Jancarik, J. & Kim, S.-H. Sparse matrix sampling. A screening method for crystallization of proteins. J. Appl. Crystallogr. 24, 409–411 (1991).
Cudney, R., Patel, S., Weisgraber, K., Newhouse, Y. & McPherson, A. Screening and optimization strategies for macromolecular crystal growth. Acta Crystallogr. D. 50, 414–423 (1994).
Mullin, J. W. W. Crystallization 4th edn. (Butterworth–Heinemann, 2001).
Asenjo, J. A. A. & Andrews, B. A. Aqueous two-phase systems for protein separation: phase separation and applications. J. Chromatogr. A 1238, 1–10 (2012).
Kuznetsov, Y. G., Malkin, A. J. & McPherson, A. The liquid protein phase in crystallization: a case study—intact immunoglobulins. J. Cryst. Growth 232, 30–39 (2001).
Chen, Q., Vekilov, P. G., Nagel, R. L. & Hirsch, R. E. Liquid–liquid phase separation in hemoglobins: distinct aggregation mechanisms of the b6 mutants. Biophys. J. 86, 1702–1712 (2004).
Chi, E. Y., Krishnan, S., Randolph, T. W. & Carpenter, J. F. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm. Res. 20, 1325–1336 (2003).
Bajaj, H. et al. Protein structural conformation and not second virial coefficient relates to long-term irreversible aggregation of a monoclonal antibody and ovalbumin in solution. Pharm. Res. 23, 1382–1394 (2006).
Nanev, C. N. On the slow kinetics of protein crystallization. Cryst. Growth Des. 7, 1533–1540 (2007).
McPherson, A. Introduction to protein crystallization. Methods 34, 254–265 (2004).
Belviso, B. D. et al. Structural characterization of the full-length anti-CD20 antibody Rituximab. Front. Mol. Biosci. 9, 823174 (2022).
Ali, I. B. et al. Assessment of blend PVDF membranes, and the effect of polymer concentration and blend composition. Membranes 8, 13 (2018).
Wu, P., Jiang, L. Y. & Hu, B. Fabrication of novel PVDF/P(VDF-co-HFP) blend hollow fiber membranes for DCMD. J. Membr. Sci. 566, 442–454 (2018).
Yi, K. et al. Diffusion coefficients of dimethyl sulfoxide (DMSO) and H2O in PAN wet spinning and its influence on morphology of nascent polyacrylonitrile (PAN) fiber. J. Eng. Fib. Fabr. 8, 107–113 (2013).
Fontananova, E., Jansen, J. C., Cristiano, A., Curcio, E. & Drioli, E. Effect of additives in the casting solution on the formation of PVDF membranes. Desalination 192, 190–197 (2006).
Lu, X. et al. Amphiphobic PVDF composite membranes for anti-fouling direct contact membrane distillation. J. Memb. Sci. 505, 61–69 (2016).
Shah, U. V., Parambil, J. V., Williams, D. R., Hinder, S. J. & Heng, J. Y. Y. Preparation and characterisation of 3D nanotemplates for protein crystallization. Powder Technol. 282, 10–18 (2015).
Bonchio, M., Carraro, M., Scorrano, G., Fontananova, E. & Drioli, E. Heterogeneous photooxidation of alcohols in water by photocatalytic membranes incorporating decatungstate. Adv. Synth. Catal. 345, 1119–1126 (2003).
Sugiyama, S. M. et al. Growth of protein crystals in hydrogels prevents osmotic shock. J. Am. Chem. Soc. 134, 5786–5789 (2012).
Caliandro, R. & Belviso, D. B. RootProf: software for multivariate analysis of unidimensional profiles. J. Appl. Cryst. 47, 1087–1096 (2014).
Acknowledgements
This work has received financial support from the European Union’s Horizon 2020 FET-OPEN research and innovation program within the AMECRYS project (http://www.amecrys-project.eu/) under grant agreement no. 712965.
Author information
Authors and Affiliations
Contributions
All authors contributed to developing this protocol and writing, refining and revising this paper. G.D.P. coordinated the AMECRYS project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Dorota Antos and Gaohong He for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Meringolo, C. et al. ACS Sustain. Chem. Eng. 6, 10069–10077 (2018): https://doi.org/10.1021/acssuschemeng.8b01407
Meringolo, C. et al. ACS Appl. Polym. Mater. 1, 326–334 (2019): https://doi.org/10.1021/acsapm.8b00105
Yang, H. et al. Crystals 9, 230 (2019): https://doi.org/10.3390/cryst9050230
Pantuso, E. et al. Sci. Rep. 10, 8902 (2020): https://doi.org/10.1038/s41598-020-65776-6
Chen, W. et al. Cryst. Growth Des. 20, 866–873 (2020): https://doi.org/10.1021/acs.cgd.9b01252
Gerard, C. J. J. et al. Cryst. Growth Des. 22, 3637–3645 (2022): https://doi.org/10.1021/acs.cgd.1c01324
Belviso, B. D. et al. Front. Mol. Biosci. 9, 823174 (2022): https://doi.org/10.3389/fmolb.2022.823174
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rajoub, N., Gerard, C.J.J., Pantuso, E. et al. A workflow for the development of template-assisted membrane crystallization downstream processing for monoclonal antibody purification. Nat Protoc 18, 2998–3049 (2023). https://doi.org/10.1038/s41596-023-00869-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00869-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.