Abstract
Human early development sets the stage for embryonic and adult life but remains difficult to investigate. A solution came from the ability of stem cells to organize into structures resembling preimplantation embryos—blastocysts—that we termed blastoids. This embryo model is available in unlimited numbers and could thus support scientific and medical advances. However, its predictive power depends on how faithfully it recapitulates the blastocyst. Here, we describe how we formed human blastoids that (1) efficiently achieve the morphology of the blastocyst and (2) form lineages according to the pace and sequence of blastocyst development, (3) ultimately forming cells that transcriptionally reflect the blastocyst (preimplantation stage). We employ three different commercially available 96- and 24-well microwell plates with results similar to our custom-made ones, and show that blastoids form in clinical in vitro fertilization medium and can be cryopreserved for shipping. Finally, we explain how blastoids replicate the directional process of implantation into endometrial organoids, specifically when these are hormonally stimulated. It takes 4 d for human blastoids to form and 10 d to prepare the endometrial implantation assay, and we have cultured blastoids up to 6 d (time-equivalent of day 13). On the basis of our experience, we anticipate that a person with ~1 year of human pluripotent stem cell culture experience and of organoid culture should be able to perform the protocol. Altogether, blastoids offer an opportunity to establish scientific and biomedical discovery programs for early pregnancy, and an ethical alternative to the use of embryos.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The main data discussed in this protocol were generated as part of the study published in the supporting primary research paper by Kagawa et al.26. Representative results obtained using this protocol are available within the article, with additional examples available from the corresponding author upon request.
References
Stamatiadis, P. et al. TEAD4 regulates trophectoderm differentiation upstream of CDX2 in a GATA3-independent manner in the human preimplantation embryo. Hum. Reprod. 37, 1760–1773 (2022).
Gerri, C. et al. Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature 587, 443–447 (2020).
Ma, H. et al. In vitro culture of cynomolgus monkey embryos beyond early gastrulation. Science 366, eaax7890 (2019).
O’Leary, T. et al. Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nat. Biotechnol. 30, 278–282 (2012).
Sunde, A. et al. Time to take human embryo culture seriously. Hum. Reprod. 31, 2174–2182 (2016).
Zinaman, M. J., Clegg, E. D., Brown, C. C., O’Connor, J. & Selevan, S. G. Estimates of human fertility and pregnancy loss. Fertil. Steril. 65, 503–509 (1996).
Jarvis, G. E. Early embryo mortality in natural human reproduction: what the data say. F1000Res. 5, 2765 (2016).
Norwitz, E. R., Schust, D. J. & Fisher, S. J. Implantation and the survival of early pregnancy. N. Engl. J. Med. 345, 1400–1408 (2001).
Wilcox, A. J., Baird, D. D. & Weinberg, C. R. Time of implantation of the conceptus and loss of pregnancy. N. Engl. J. Med. 340, 1796–1799 (1999).
Gluckman, P. D., Hanson, M. A., Cooper, C. & Thornburg, K. L. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 359, 61–73 (2008).
Barker, D. J. et al. Fetal nutrition and cardiovascular disease in adult life. Lancet 341, 938–941 (1993).
Cha, J., Sun, X. & Dey, S. K. Mechanisms of implantation: strategies for successful pregnancy. Nat. Med. 18, 1754–1767 (2012).
Siriwardena, D. & Boroviak, T. E. Evolutionary divergence of embryo implantation in primates. Philos. Trans. R. Soc. Lond. B 377, 20210256 (2022).
Fu, J., Warmflash, A. & Lutolf, M. P. Stem-cell-based embryo models for fundamental research and translation. Nat. Mater. 20, 132–144 (2021).
Rivron, N. C. et al. Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111 (2018).
Amita, M. et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc. Natl Acad. Sci. USA 110, E1212–E1221 (2013).
Cinkornpumin, J. K. et al. Naive human embryonic stem cells can give rise to cells with a trophoblast-like transcriptome and methylome. Stem Cell Rep. 15, 198–213 (2020).
Castel, G. et al. Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Rep. 33, 108419 (2020).
Io, S. et al. Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell 28, 1023–1039.e13 (2021).
Guo, G. et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 28, 1040–1056.e6 (2021).
Yanagida, A. et al. Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell 28, 1016–1022.e4 (2021).
Okae, H. et al. Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018).
Meistermann, D. et al. Integrated pseudotime analysis of human pre-implantation embryo single-cell transcriptomes reveals the dynamics of lineage specification. Cell Stem Cell 28, 1625–1640.e6 (2021).
Radley, A., Corujo-Simon, E., Nichols, J., Smith, A. & Dunn, S.-J. Entropy sorting of single-cell RNA sequencing data reveals the inner cell mass in the human pre-implantation embryo. Stem Cell Rep. 18, 47–63 (2022).
Seong, J. et al. Epiblast inducers capture mouse trophectoderm stem cells in vitro and pattern blastoids for implantation in utero. Cell Stem Cell 29, 1102–1118.e8 (2022).
Kagawa, H. et al. Human blastoids model blastocyst development and implantation. Nature 610, 600–605 (2022).
Li, R., Zhong, C. & Izpisua Belmonte, J. C. Time matters: human blastoids resemble the sequence of blastocyst development. Cell 185, 581–584 (2022).
Nichols, J. & Smith, A. Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 (2009).
Taubenschmid-Stowers, J. et al. 8C-like cells capture the human zygotic genome activation program in vitro. Cell Stem Cell 29, 449–459.e6 (2022).
Mazid, M. A. et al. Rolling back human pluripotent stem cells to an eight-cell embryo-like stage. Nature 605, 315–324 (2022).
Posfai, E. et al. Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 23, 49–60 (2021).
Guo, G. et al. Epigenetic resetting of human pluripotency. Development 144, 2748–2763 (2017).
Takashima, Y. et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269 (2014).
Stirparo, G. G. et al. Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast. Development 145, dev158501 (2018).
Theunissen, T. W. et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19, 502–515 (2016).
Bayerl, J. et al. Principles of signaling pathway modulation for enhancing human naive pluripotency induction. Cell Stem Cell 28, 1549–1565.e12 (2021).
Paepe, C. D. et al. Human trophectoderm cells are not yet committed. Hum. Reprod. 28, 740–749 (2013).
Slack, J. M. W. & Slack. From Egg to Embryo: Regional Specification in Early Development. (Cambridge University Press, 1991).
Zijlmans, D. W. et al. Integrated multi-omics reveal polycomb repressive complex 2 restricts human trophoblast induction. Nat. Cell Biol. 24, 858–871 (2022).
Kumar, B. et al. Polycomb repressive complex 2 shields naïve human pluripotent cells from trophectoderm differentiation. Nat. Cell Biol. 24, 845–857 (2022).
Yang, Y. et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 169, 243–257.e25 (2017).
Nakamura, T. et al. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537, 57–62 (2016).
Bredenkamp, N. et al. Wnt inhibition facilitates RNA-mediated reprogramming of human somatic cells to naive pluripotency. Stem Cell Rep. 13, 1083–1098 (2019).
Guo, G. et al. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep. 6, 437–446 (2016).
Khan, S. A. et al. Probing the signaling requirements for naive human pluripotency by high-throughput chemical screening. Cell Rep. 35, 109233 (2021).
Kagawa, H. et al. Protocol for human blastoids modeling blastocyst development and implantation. J. Vis. Exp. https://doi.org/10.3791/63388 (2022).
Stephenson, R. O., Rossant, J. & Tam, P. P. L. Intercellular interactions, position, and polarity in establishing blastocyst cell lineages and embryonic axes. Cold Spring Harb. Perspect. Biol. 4, a008235 (2012).
Korotkevich, E. et al. The apical domain is required and sufficient for the first lineage segregation in the mouse embryo. Dev. Cell 40, 235–247.e7 (2017).
Kim, E. J. Y., Korotkevich, E. & Hiiragi, T. Coordination of cell polarity, mechanics and fate in tissue self-organization. Trends Cell Biol. 28, 541–550 (2018).
Hirate, Y. et al. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 23, 1181–1194 (2013).
Cockburn, K., Biechele, S., Garner, J. & Rossant, J. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 23, 1195–1201 (2013).
Nishioka, N. et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 (2009).
Yu, F.-X. et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 (2012).
Rivron, N. C. et al. Tissue deformation spatially modulates VEGF signaling and angiogenesis. Proc. Natl Acad. Sci. USA 109, 6886–6891 (2012).
Vrij, E. et al. Directed assembly and development of material-free tissues with complex architectures. Adv. Mater. 28, 4032–4039 (2016).
Frias-Aldeguer, J. et al. Embryonic signals perpetuate polar-like trophoblast stem cells and pattern the blastocyst axis. Preprintat at bioRxiv https://doi.org/10.1101/510362 (2020).
Rostovskaya, M., Andrews, S., Reik, W. & Rugg-Gunn, P. J. Amniogenesis occurs in two independent waves in primates. Cell Stem Cell 29, 744–759.e6 (2022).
Ohgushi, M. & Sasai, Y. Lonely death dance of human pluripotent stem cells: ROCKing between metastable cell states. Trends Cell Biol. 21, 274–282 (2011).
Pham, T. X. A. et al. Modeling human extraembryonic mesoderm cells using naive pluripotent stem cells. Cell Stem Cell 29, 1346–1365.e10 (2022).
Zhao, C. et al. Reprogrammed iBlastoids contain amnion-like cells but not trophectoderm. Preprint at bioRxiv https://doi.org/10.1101/2021.05.07.442980 (2021).
Clark, A. T. et al. Human embryo research, stem cell-derived embryo models and in vitro gametogenesis: Considerations leading to the revised ISSCR guidelines. Stem Cell Rep. 16, 1416–1424 (2021).
Lovell-Badge, R. et al. ISSCR guidelines for stem cell research and clinical translation: the 2021 update. Stem Cell Rep. 16, 1398–1408 (2021).
Singh, H., Nardo, L., Kimber, S. J. & Aplin, J. D. Early stages of implantation as revealed by an in vitro model. Reproduction 139, 905–914 (2010).
Heng, S. et al. Podocalyxin inhibits human embryo implantation in vitro and luminal podocalyxin in putative receptive endometrium is associated with implantation failure in fertility treatment. Fertil. Steril. 116, 1391–1401 (2021).
Hannan, N. J., Paiva, P., Dimitriadis, E. & Salamonsen, L. A. Models for study of human embryo implantation: choice of cell lines? Biol. Reprod. 82, 235–245 (2010).
Zhang, D. et al. A new model for embryo implantation: coculture of blastocysts and Ishikawa cells. Gynecol. Endocrinol. 28, 288–292 (2012).
Ruane, P. T. et al. The effects of hyaluronate-containing medium on human embryo attachment to endometrial epithelial cells. Hum. Reprod. Open 2020, hoz033 (2020).
Turco, M. Y. et al. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 19, 568–577 (2017).
Boretto, M. et al. Development of organoids from mouse and human endometrium showing endometrial epithelium physiology and long-term expandability. Development 144, 1775–1786 (2017).
Yu, S.-L. et al. Transcriptomic analysis and competing endogenous RNA network in the human endometrium between proliferative and mid-secretory phases. Exp. Ther. Med. 21, 660 (2021).
Wang, W. et al. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat. Med. 26, 1644–1653 (2020).
Tulac, S. et al. Dickkopf-1, an inhibitor of Wnt signaling, is regulated by progesterone in human endometrial stromal cells. J. Clin. Endocrinol. Metab. 91, 1453–1461 (2006).
Garcia-Alonso, L. et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat. Genet. 53, 1698–1711 (2021).
Vollset, S. E. et al. Fertility, mortality, migration, and population scenarios for 195 countries and territories from 2017 to 2100: a forecasting analysis for the Global Burden of Disease Study. Lancet 396, 1285–1306 (2020).
Marx, V. Modeling the early embryo. Nat. Methods 19, 644–648 (2022).
Dumortier, J. G. et al. Hydraulic fracturing and active coarsening position the lumen of the mouse blastocyst. Science 365, 465–468 (2019).
Salmen, F. et al. High-throughput total RNA sequencing in single cells using VASA-seq. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01361-8 (2022).
Clark, S. J. et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018).
Fan, Y. et al. Generation of human blastocyst-like structures from pluripotent stem cells. Cell Discov. 7, 81 (2021).
Liu, X. et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 591, 627–632 (2021).
Sozen, B. et al. Reconstructing aspects of human embryogenesis with pluripotent stem cells. Nat. Commun. 12, 5550 (2021).
Yu, L. et al. Blastocyst-like structures generated from human pluripotent stem cells. Nature 591, 620–626 (2021).
Chen, Y., Siriwardena, D., Penfold, C., Pavlinek, A. & Boroviak, T. E. An integrated atlas of human placental development delineates essential regulators of trophoblast stem cells. Development 149, dev200171 (2022).
Shapiro, B. S., Daneshmand, S. T., Garner, F. C., Aguirre, M. & Thomas, S. Large blastocyst diameter, early blastulation, and low preovulatory serum progesterone are dominant predictors of clinical pregnancy in fresh autologous cycles. Fertil. Steril. 90, 302–309 (2008).
Baumann, K. A role model of human blastocysts. Nat. Rev. Mol. Cell Biol. 23, 91 (2022).
Posfai, E., Lanner, F., Mulas, C. & Leitch, H. G. All models are wrong, but some are useful: establishing standards for stem cell-based embryo models. Stem Cell Rep. 16, 1117–1141 (2021).
Hyun, I., Munsie, M., Pera, M. F., Rivron, N. C. & Rossant, J. Toward guidelines for research on human embryo models formed from stem cells. Stem Cell Rep. 14, 169–174 (2020).
Collier, A. J. et al. Comprehensive cell surface protein profiling identifies specific markers of human naive and primed pluripotent states. Cell Stem Cell 20, 874–890.e7 (2017).
Bredenkamp, N., Stirparo, G. G., Nichols, J., Smith, A. & Guo, G. The cell-surface marker sushi containing domain 2 facilitates establishment of human naive pluripotent stem cells. Stem Cell Rep. 12, 1212–1222 (2019).
Baker, D. & Barbaric, I. Characterizing the genetic stability of human naïve and primed pluripotent stem cells. Methods Mol. Biol. 2416, 267–284 (2022).
Boretto, M. et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 21, 1041–1051 (2019).
Fujii, M., Matano, M., Nanki, K. & Sato, T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 10, 1474–1485 (2015).
Shin, W. & Kim, H. J. 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert. Nat. Protoc. 17, 910–939 (2022).
Heijmans, J. et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 3, 1128–1139 (2013).
Rugg-Gunn, P. J. Induction of human naïve pluripotency using chemical resetting. Methods Mol. Biol. 2416, 29–37 (2022).
Behringer, R., Gertsenstein, M., Nagy, K. V. & Nagy, A. Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2013).
Jeschke, U. et al. The human endometrium expresses the glycoprotein mucin-1 and shows positive correlation for Thomsen-Friedenreich epitope expression and galectin-1 binding. J. Histochem. Cytochem. 57, 871–881 (2009).
Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC-Co grant agreement no. 101002317 ‘BLASTOID: a discovery platform for early human embryogenesis’). This project has received funding from the Austrian Science Fund (FWF), Lise Meitner Programme M3131-B, H.H.K. is supported by FWF. This project has received funding from the European Union’s Framework Programme for Research and Innovation Horizon 2020 (2014–2020) under the Marie Skłodowska-Curi grant agreement no. 101026451. H.K. is supported by the Japan Society for the Promotion of Science Overseas Research Fellowships. G.S. is supported by the HFSP number RGY0081/2019. We thank Y. Takashima for sharing the H9 and H9-GFP cell lines; A. Smith, P. Andrews and G. Guo for sharing the HNES1, Shef6, niPSC 16.2b and cR-NCRM2 cell lines; H. Baharvand for sharing the endometrial organoids; S. Srinivas for sharing the scRNA-seq data of peri-gastrulation embryo; A. Bykov and L. Cochella for technical assistance for SMARTSeq2 library preparation; and the NGS, Biooptic and Stem Cell facility at IMBA for critical assistance.
Author information
Authors and Affiliations
Contributions
H.H.K., A.J., H.K. and N.R. conceived the study; N.R. supervised the project; H.K., A.J., H.H.K., T.M.S. and N.R. designed the blastoid experiments; H.K., A.J., H.H.K., T.M.S., J.S. and Y.S.o.R. performed blastoid experiments; G.S., L.D. and M.N. performed the bioinformatic analysis of scRNA-seq datasets; H.K., A.J., H.H.K., T.M.S., Y.S.o.R. and N.R. analyzed data; N.R. wrote the manuscript with help from all the authors.
Corresponding author
Ethics declarations
Competing interests
The Institute for Molecular Biotechnology, Austrian Academy of Sciences has filed patent application EP21151455.9 describing the protocols for human blastoid formation and for the blastoid–endometrium interaction assay. H.K., A.J., H.H.K. and N.R. are the inventors on this patent. All other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Susana Chuva de Sousa Lopes, Kiichiro Tomoda and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key paper using this protocol
Kagawa, H. et al. Nature 601, 600–605 (2022): https://doi.org/10.1038/s41586-021-04267-8
Extended data
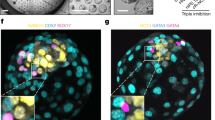
Extended Data Fig. 1 Triple inhibition of Hippo, ERK and TGFβ pathways leads to efficient and robust formation of human blastoids.
Time course bright-field images of PXGL hPSCs aggregates and blastoid formation within AggreWell (top) and microwell arrays (bottom) in PALLY medium. Scale bars, 400 μm.
Extended Data Fig. 2 Human blastoids formation in IVF medium and vitrification.
a, Bright-field image of human blastoids formed after 48 h stimulation with PALLY medium followed by the use of IVF medium (G2, Vitrolife) for the last 2 d. Scale bars, 400 µm. b, Bright-field image of control (top) and vitrified-thawed human blastoid (bottom) and after 2 d extended culture on Matrigel-coated plate. Scale bars, 100 µm. c, Confocal immunofluorescence image of OCT4 (yellow) and aPKC (gray) in control (top) and vitrified-thawed human blastoid (bottom) cultured on Matrigel-coated plate for 2 d, counterstained with phalloidin marking F-actin (cyan). Arrows point to the pro-amniotic-like cavity. Scale bar, 100 μm.
Supplementary information
Supplementary Information
Supplementary Tables 1and 2.
Supplementary Video 1
Human blastoids attach via the polar trophectoderm. Live imaging of human blastoids attached via the polar region to hormonally stimulated layers of endometrial epithelial cells and challenged by pipetting liquid in their vicinity.
Supplementary Video 2
Human blastoids on non-hormonally stimulated endometrial layers. Live imaging of human blastoids on non-hormonally stimulated layers of endometrial epithelial cells, and challenged by pipetting liquid in their vicinity. Blastoids do not show attachment.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Heidari Khoei, H., Javali, A., Kagawa, H. et al. Generating human blastoids modeling blastocyst-stage embryos and implantation. Nat Protoc 18, 1584–1620 (2023). https://doi.org/10.1038/s41596-023-00802-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00802-1
This article is cited by
-
A combined clinical and specific genes’ model to predict live birth for in vitro fertilization and embryo transfer patients
BMC Pregnancy and Childbirth (2023)
-
Combining Endometrial Assembloids and Blastoids to Delineate the Molecular Roadmap of Implantation
Stem Cell Reviews and Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.