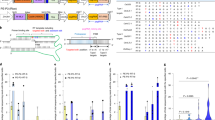
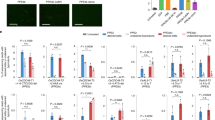
Abstract
Prime editors (PEs), which can install desired base edits without donor DNA or double-strand breaks, have been used in plants and can, in principle, accelerate crop improvement and breeding. However, their editing efficiency in plants is generally low. Optimizing the prime editing guide RNA (pegRNA) by designing the sequence on the basis of melting temperature, using dual-pegRNAs and engineering PEs have all been shown to enhance PE efficiency. In addition, an automated pegRNA design platform, PlantPegDesigner, has been developed on the basis of rice prime editing experimental data. In this protocol, we present detailed protocols for designing and optimizing pegRNAs using PlantPegDesigner, constructing engineered plant PE vectors with enhanced editing efficiency for prime editing, evaluating prime editing efficiencies using a reporter system and comparing the effectiveness and byproducts of PEs by deep amplicon sequencing. Using this protocol, researchers can construct optimized pegRNAs for prime editing in 4–7 d and obtain prime-edited rice or wheat plants within 3 months.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All the data shown in this protocol are deposited in NCBI BioProject under accession codes PRJNA605069, PRJNA605074, and PRJNA702010. An example dataset for amplicon deep sequencing analysis used in this protocol is available on GitHub at https://github.com/ReiGao/GEanalysis.
Code availability
All the code used in this protocol is available on GitHub at https://github.com/ReiGao/GEanalysis. The PlantPegDesigner web application code is available at https://github.com/JinShuai001/PlantPegDesigner.
References
Gao, C. Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635 (2021).
Li, Y., Li, W. & Li, J. The CRISPR/Cas9 revolution continues: from base editing to prime editing in plant science. J. Genet. Genomics 48, 661–670 (2021).
Li, G., Liu, Y. G. & Chen, Y. Genome-editing technologies: the gap between application and policy. Sci. China Life Sci. 62, 1534–1538 (2019).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Lin, Q. et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585 (2020).
Yang, L., Yang, B. & Chen, J. One prime for all editing. Cell 179, 1448–1450 (2019).
Jin, S. et al. Genome-wide specificity of prime editors in plants. Nat. Biotechnol. 39, 1292–1299 (2021).
Gao, R. et al. Genomic and transcriptomic analyses of prime editing guide RNA-independent off-target effects by prime editors. CRISPR J. 5, 276–293 (2022).
Kim, D. Y., Moon, S. B., Ko, J. H., Kim, Y. S. & Kim, D. Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res. 48, 10576–10589 (2020).
Zong, Y. et al. An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01254-w (2022).
Song, M. et al. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nat. Commun. 12, 5617 (2021).
Park, S. J. et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol. 22, 170 (2021).
Liu, P. et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. 12, 2121 (2021).
Velimirovic, M. et al. Peptide fusion improves prime editing efficiency. Nat. Commun. 13, 3512 (2022).
Jiang, Y. et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes of maize. Genome Biol. 21, 257 (2020).
Liu, Y. et al. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res. 31, 1134–1136 (2021).
Nelson, J. W. et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 40, 402–410 (2022).
Chai, Y. et al. MS2 RNA aptamer enhances prime editing in rice. Preprint at bioRxiv https://doi.org/10.1101/2021.10.20.465209 (2021).
Chen, P. J. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e29 (2021).
Ferreira da Silva, J. F. et al. Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair. Nat. Commun. 13, 760 (2022).
Anzalone, A. V. et al. Programmable large DNA deletion, replacement, integration, and inversion with twin prime editing and site-specific recombinases. Nat. Biotechnol. 40, 731–740 (2021).
Ioannidi, E. I. et al. Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases. Preprint at bioRxiv https://doi.org/10.1101/2021.11.01.466786 (2021).
Kim, H. K. et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol. 39, 198–206 (2020).
Liu, Y. et al. Efficient generation of mouse models with the prime editing system. Cell Discov. 6, 27 (2020).
Lin, Q. et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 39, 923–927 (2021).
Standage-Beier, K., Tekel, S. J., Brafman, D. A. & Wang, X. Prime editing guide RNA design automation using PINE-CONE. ACS Synth. Biol. 10, 422–427 (2021).
Bhagwat, A. M. et al. Multicrispr: gRNA design for prime editing and parallel targeting of thousands of targets. Life Sci. Alliance 3, e202000757 (2020).
Chow, R. D., Chen, J. S., Shen, J. & Chen, S. A web tool for the design of prime-editing guide RNAs. Nat. Biomed. Eng. 5, 190–194 (2020).
Hsu, J. Y. et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat. Commun. 12, 1034 (2021).
Morris, J.A., Rahman, J.A., Guo, X. & Sanjana, N.E. Automated design of CRISPR prime editors for 56,000 human pathogenic variants. iScience 24, 103380 (2021).
Siegner, S. M., Karasu, M. E., Schröder, M. S., Kontarakis, Z. & Corn, J. E. PnB Designer: a web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinformatics 22, 101 (2021).
Hwang, G. H. et al. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Res. 49, W499–W504 (2021).
Anderson, M. V., Haldrup, J., Thomsen, E. A., Wolff, J. H. & Mikkelsen, J. G. pegIT—a web-based design tool for prime editing. Nucleic Acids Res. 49, W505–W509 (2021).
Li, Y., Chen, J., Tsai, S. Q. & Cheng, Y. Easy-Prime: a machine learning-based prime editor design tool. Genome Biol. 22, 235 (2021).
Choi, J. et al. Precise genomic deletions using paired prime editing. Nat. Biotechnol. 40, 218–226 (2022).
Jiang, T., Zhang, X. O., Weng, Z. & Xue, W. Deletion and replacement of long genomic sequences using prime editing. Nat. Biotechnol. 40, 227–234 (2022).
Zhuang, Y. et al. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nat. Chem. Biol. 18, 29–37 (2022).
Bae, S., Park, J. & Kim, J. S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Singh, R., Kuscu, C., Quinlan, A., Qi, Y. & Adli, M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 43, e118 (2015).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Kim, N. et al. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat. Biotechnol. 38, 1328–1336 (2020).
Wang, D. et al. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat. Commun. 10, 4284 (2019).
Labuhn, M. et al. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR–Cas9 applications. Nucleic Acids Res. 46, 1375–1385 (2018).
Tang, X. et al. Plant prime editors enable precise gene editing in rice cells. Mol. Plant 13, 667–670 (2020).
Li, H., Li, J., Chen, J., Yan, L. & Xia, L. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 13, 671–674 (2020).
Xu, W. et al. Versatile nucleotides substitution in plant using an improved prime editing system. Mol. Plant 13, 675–678 (2020).
Xu, R. et al. Development of plant prime-editing systems for precise genome editing. Plant Commun. 1, 100043 (2020).
Hua, K., Jiang, Y., Tao, X. & Zhu, J. K. Precision genome engineering in rice using prime editing system. Plant Biotechnol. J. 18, 2167–2169 (2020).
Butt, H. et al. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 18, 2370–2372 (2020).
Wang, L. et al. Spelling changes and fluorescent tagging with prime editing vectors for plants. Front. Genome Ed. 3, 617553 (2021).
Shan, Q., Wang, Y., Li, J. & Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410 (2014).
Liu, Z. et al. Precise editing of methylated cytosine in Arabidopsis thaliana using a human APOBEC3Bctd–Cas9 fusion. Sci. China Life Sci. 65, 219–222 (2021).
Tang, S. et al. Targeted DNA demethylation produces heritable epialleles in rice. Sci. China Life Sci. 65, 753–756 (2021).
Lin, Q. et al. Genome editing in plants with MAD7 nuclease. J. Genet. Genomics 48, 444–451 (2021).
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S. & Thompson, W. F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325 (2006).
Liang, Z. et al. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13, 413–430 (2018).
Jin, S., Gao, Q. & Gao, C. An unbiased method for evaluating the genome-wide specificity of base editors in rice. Nat. Protoc. 16, 431–457 (2021).
Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019).
Jin, S. et al. Rationally designed APOBEC3B cytosine base editors with improved specificity. Mol. Cell 79, 728–740 (2020).
Acknowledgements
We thank K. T. Zhao for his insightful comments on the manuscript. This work was supported by the Ministry of Agriculture and Rural Affairs of China, the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24020102 to C.G.), the Young Elite Scientists Sponsorship Program of the China Association for Science and Technology (2020QNRC001 to S.J.) and the Postdoctoral Innovative Talent Support Program of China (BX2021353 to Q.L.).
Author information
Authors and Affiliations
Contributions
C.G. S.J., Q.L. and Q.G. wrote the manuscript; Q.L. and S.J. designed figures; C.G. supervised the project;
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Goetz Hensel, Yiping Qi and Seiichi Toki for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Lin, Q. et al. Nat. Biotechnol. 38, 582–585 (2020): https://doi.org/10.1038/s41587-020-0455-x
Lin, Q. et al. Nat. Biotechnol. 39, 923–927 (2021): https://doi.org/10.1038/s41587-021-00868-w
Zong, Y. et al. Nat. Biotechnol. 40, 1394–1402 (2022): https://doi.org/10.1038/s41587-022-01254-w
Extended data
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Notes 1–3.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jin, S., Lin, Q., Gao, Q. et al. Optimized prime editing in monocot plants using PlantPegDesigner and engineered plant prime editors (ePPEs). Nat Protoc 18, 831–853 (2023). https://doi.org/10.1038/s41596-022-00773-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00773-9
This article is cited by
-
Application of genome editing techniques to regulate gene expression in crops
BMC Plant Biology (2024)
-
Mapping nucleosome-resolution chromatin organization and enhancer-promoter loops in plants using Micro-C-XL
Nature Communications (2024)
-
Strand-preferred base editing of organellar and nuclear genomes using CyDENT
Nature Biotechnology (2023)
-
Tuning plant phenotypes by precise, graded downregulation of gene expression
Nature Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.