Abstract
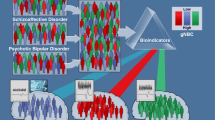
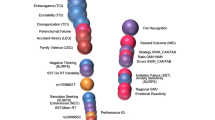
Normative modeling is an emerging and innovative framework for mapping individual differences at the level of a single subject or observation in relation to a reference model. It involves charting centiles of variation across a population in terms of mappings between biology and behavior, which can then be used to make statistical inferences at the level of the individual. The fields of computational psychiatry and clinical neuroscience have been slow to transition away from patient versus ‘healthy’ control analytic approaches, probably owing to a lack of tools designed to properly model biological heterogeneity of mental disorders. Normative modeling provides a solution to address this issue and moves analysis away from case–control comparisons that rely on potentially noisy clinical labels. Here we define a standardized protocol to guide users through, from start to finish, normative modeling analysis using the Predictive Clinical Neuroscience toolkit (PCNtoolkit). We describe the input data selection process, provide intuition behind the various modeling choices and conclude by demonstrating several examples of downstream analyses that the normative model may facilitate, such as stratification of high-risk individuals, subtyping and behavioral predictive modeling. The protocol takes ~1–3 h to complete.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data used in this protocol are available on GitHub (https://github.com/predictive-clinical-neuroscience/PCNtoolkit-demo/tree/main/tutorials/BLR_protocol) and Zenodo86 (https://zenodo.org/record/5592153#.YjL7PY_P2UI) in csv. files. We also include a template csv. file to help format user’s own data into the correct form for running the protocol using their own dataset.
Code availability
All code is available on GitHub (https://github.com/predictive-clinical-neuroscience/PCNtoolkit-demo/tree/main/tutorials/BLR_protocol) in the format of Python notebooks that can be run in the cloud (for free) using Google Colab (https://colab.research.google.com/github/predictive-clinical-neuroscience/PCNtoolkit-demo/blob/main/tutorials/BLR_protocol/BLR_normativemodel_protocol.ipynb). We have also shared the GitHub repository on Zenodo (https://zenodo.org/record/5592153#.YjL7PY_P2UI) to create a citable DOI for this software that also allows versions that are necessary as additional code and tutorials may be added over time86.
References
Wang, D. et al. Parcellating cortical functional networks in individuals. Nat. Neurosci. 18, 1853–1860 (2015).
Finn, E. S. & Constable, R. T. Individual variation in functional brain connectivity: implications for personalized approaches to psychiatric disease. Dialogues Clin. Neurosci. 18, 277–287 (2016).
Braga, R. M. & Buckner, R. L. Parallel interdigitated distributed networks within the individual estimated by intrinsic functional connectivity. Neuron 95, 457–471 (2017).
Poldrack, R. A. Precision neuroscience: dense sampling of individual brains. Neuron 95, 727–729 (2017).
Vanderwal, T. et al. Individual differences in functional connectivity during naturalistic viewing conditions. Neuroimage 157, 521–530 (2017).
Braun, U. et al. From maps to multi-dimensional network mechanisms of mental disorders. Neuron 97, 14–31 (2018).
Gratton, C. et al. Defining individual-specific functional neuroanatomy for precision psychiatry. Biol. Psychiatry 88, 28–39 (2020).
Hyman, S. E. Can neuroscience be integrated into the DSM-V? Nat. Rev. Neurosci. 8, 725–732 (2007).
Insel, T. et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am. J. Psychiatry 167, 748–751 (2010).
Michelini, G., Palumbo, I. M., DeYoung, C. G., Latzman, R. D. & Kotov, R. Linking RDoC and HiTOP: a new interface for advancing psychiatric nosology and neuroscience. Clin. Psychol. Rev. 86, 102025 (2021).
Narrow, W. E. & Kuhl, E. A. Dimensional approaches to psychiatric diagnosis in DSM-5. J. Ment. Health Policy Econ. 14, 197–200 (2011).
Cuthbert, B. N. & Insel, T. R. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 11, 126 (2013).
Sanislow, C. A. RDoC at 10: changing the discourse for psychopathology. World Psychiatry 19, 311–312 (2020).
Kotov, R. et al. The Hierarchical Taxonomy of Psychopathology (HiTOP): a dimensional alternative to traditional nosologies. J. Abnorm. Psychol. 126, 454–477 (2017).
Kotov, R. et al. The Hierarchical Taxonomy of Psychopathology (HiTOP): a quantitative nosology based on consensus of evidence. Annu. Rev. Clin. Psychol. 17, 081219–093304 (2021).
Haro, J. M. et al. ROAMER: roadmap for mental health research in Europe. Int. J. Methods Psychiatr. Res. 23, 1–14 (2014).
Schumann, G. et al. Stratified medicine for mental disorders. Eur. Neuropsychopharmacol. 24, 5–50 (2014).
Feczko, E. et al. The heterogeneity problem: approaches to identify psychiatric subtypes. Trends Cogn. Sci. 23, 584–601 (2019).
Shen, X. et al. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat. Protoc. 12, 506–518 (2017).
Sripada, C. et al. Basic units of inter-individual variation in resting state connectomes. Sci. Rep. 9, 1900 (2019).
Woo, C.-W., Chang, L. J., Lindquist, M. A. & Wager, T. D. Building better biomarkers: brain models in translational neuroimaging. Nat. Neurosci. 20, 365–377 (2017).
Marquand, A. F. et al. Conceptualizing mental disorders as deviations from normative functioning. Mol. Psychiatry 24, 1415–1424 (2019).
Gau, R. et al. Brainhack: developing a culture of open, inclusive, community-driven neuroscience. Neuron 109, 1769–1775 (2021).
Olah, C. & Carter, S. Research debt. Distill 2, e5 (2017).
Fraza, C. J., Dinga, R., Beckmann, C. F. & Marquand, A. F. Warped Bayesian linear regression for normative modelling of big data. Neuroimage 245, 118715 (2021).
Dinga, R. et al. Normative modeling of neuroimaging data using generalized additive models of location scale and shape. Preprint at bioRxiv https://doi.org/10.1101/2021.06.14.448106 (2021).
Kia, S. M. et al. Hierarchical Bayesian regression for multi-site normative modeling of neuroimaging data. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 (eds. Martel, A. L. et al.) 699–709 (Springer International, 2020); https://doi.org/10.1007/978-3-030-59728-3_68
Kia, S. M. et al. Federated multi-site normative modeling using hierarchical Bayesian regression. Preprint at bioRxiv https://doi.org/10.1101/2021.05.28.446120 (2021).
Floris, D. L. et al. Atypical brain asymmetry in autism—a candidate for clinically meaningful stratification. Biol. Psychiatry Cogn. Neurosci. Neuroimaging https://doi.org/10.1016/j.bpsc.2020.08.008 (2020).
Zabihi, M. et al. Dissecting the heterogeneous cortical anatomy of autism spectrum disorder using normative models. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 4, 567–578 (2019).
Zabihi, M. et al. Fractionating autism based on neuroanatomical normative modeling. Transl. Psychiatry 10, 1–10 (2020).
Wolfers, T. et al. Individual differences v. the average patient: mapping the heterogeneity in ADHD using normative models. Psychol. Med. 50, 314–323 (2020).
Wolfers, T. et al. Refinement by integration: aggregated effects of multimodal imaging markers on adult ADHD. J. Psychiatry Neurosci. 42, 386–394 (2017).
Verdi, S., Marquand, A. F., Schott, J. M. & Cole, J. H. Beyond the average patient: how neuroimaging models can address heterogeneity in dementia. Brain https://doi.org/10.1093/brain/awab165 (2021).
Wolfers, T. et al. Replicating extensive brain structural heterogeneity in individuals with schizophrenia and bipolar disorder. Human Brain Mapp. https://doi.org/10.1002/hbm.25386 (2020).
Wolfers, T. et al. Mapping the heterogeneous phenotype of schizophrenia and bipolar disorder using normative models. JAMA Psychiatry 75, 1146–1155 (2018).
Wolfers, T. et al. Replicating extensive brain structural heterogeneity in individuals with schizophrenia and bipolar disorder. Hum. Brain Mapp. 42, 2546–2555 (2021).
Sripada, C., Angstadt, M., Rutherford, S. & Taxali, A. Brain network mechanisms of general intelligence. Preprint at bioRxiv https://doi.org/10.1101/657205 (2019).
Sripada, C. et al. Brain Connectivity Patterns in Children Linked to Neurocognitive Abilities. Preprint at bioRxiv https://doi.org/10.1101/2020.09.10.291500 (2020).
Rosenberg, M. D. et al. A neuromarker of sustained attention from whole-brain functional connectivity. Nat. Neurosci. 19, 165–171 (2015).
Rosenberg, M. D. et al. Functional connectivity predicts changes in attention observed across minutes, days, and months. Proc. Natl Acad. Sci. USA 117, 3797–3807 (2020).
Marquand, A. F., Haak, K. V. & Beckmann, C. F. Functional corticostriatal connection topographies predict goal directed behaviour in humans. Nat. Hum. Behav. 1, 0146 (2017).
Marquand, A. et al. Quantitative prediction of subjective pain intensity from whole-brain fMRI data using Gaussian processes. Neuroimage 49, 2178–2189 (2010).
Wager, T. D. et al. An fMRI-based neurologic signature of physical pain. N. Engl. J. Med. 368, 1388–1397 (2013).
Sripada, C., Angstadt, M., Rutherford, S., Taxali, A. & Shedden, K. Toward a “treadmill test” for cognition: improved prediction of general cognitive ability from the task activated brain. Hum. Brain Mapp. 41, 3186–3197 (2020).
Taxali, A., Angstadt, M., Rutherford, S. & Sripada, C. Boost in test–retest reliability in resting state fMRI with predictive modeling. Cereb. Cortex 31, 2822–2833 (2021).
Finn, E. S. et al. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci. 18, 1664–1671 (2015).
Wang, H.-T. et al. Finding the needle in a high-dimensional haystack: canonical correlation analysis for neuroscientists. Neuroimage 216, 116745 (2020).
Smith, S. M. et al. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat. Neurosci. 18, 1565–1567 (2015).
Dadi, K. et al. Benchmarking functional connectome-based predictive models for resting-state fMRI. Neuroimage 192, 115–134 (2019).
Lake, E. M. R. et al. The functional brain organization of an individual allows prediction of measures of social abilities transdiagnostically in autism and attention-deficit/hyperactivity disorder. Biol. Psychiatry 86, 315–326 (2019).
Cole, J. H. & Franke, K. Predicting age using neuroimaging: innovative brain ageing biomarkers. Trends Neurosci. 40, 681–690 (2017).
Han, L. K. M. et al. Brain aging in major depressive disorder: results from the ENIGMA major depressive disorder working group. Mol. Psychiatry 26, 5124–5139 (2021).
Sturmfels, P. et al. A domain guided CNN architecture for predicting age from structural brain images. Preprint at arXiv https://doi.org/10.48550/arXiv.1808.04362 (2018).
Alfaro-Almagro, F. et al. Image processing and quality control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 166, 400–424 (2018).
Donders, A. R. T., van der Heijden, G. J. M. G., Stijnen, T. & Moons, K. G. M. Review: a gentle introduction to imputation of missing values. J. Clin. Epidemiol. 59, 1087–1091 (2006).
Madley-Dowd, P., Hughes, R., Tilling, K. & Heron, J. The proportion of missing data should not be used to guide decisions on multiple imputation. J. Clin. Epidemiol. 110, 63–73 (2019).
Burt, J. B., Helmer, M., Shinn, M., Anticevic, A. & Murray, J. D. Generative modeling of brain maps with spatial autocorrelation. Neuroimage 220, 117038 (2020).
Shinn, M. et al. Spatial and temporal autocorrelation weave human brain networks. Preprint at bioRxiv https://doi.org/10.1101/2021.06.01.446561 (2021).
Smith, S. M. & Nichols, T. E. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage 44, 83–98 (2009).
Guo, C., Kang, J. & Johnson, T. D. A spatial Bayesian latent factor model for image-on-image regression. Biometrics https://doi.org/10.1111/biom.13420 (2020).
Woolrich, M. W., Behrens, T. E. J. & Smith, S. M. Constrained linear basis sets for HRF modelling using variational Bayes. Neuroimage 21, 1748–1761 (2004).
Liu, W., Zhu, P., Anderson, J. S., Yurgelun-Todd, D. & Fletcher, P. T. Spatial regularization of functional connectivity using high-dimensional Markov random fields. Med. Image Comput. Comput. Assist. Interv. 13, 363–370 (2010).
Song, H. F., Kennedy, H. & Wang, X.-J. Spatial embedding of structural similarity in the cerebral cortex. Proc. Natl Acad. Sci. USA 111, 16580–16585 (2014).
Roberts, J. A. et al. The contribution of geometry to the human connectome. Neuroimage 124, 379–393 (2016).
Bijsterbosch, J. et al. Challenges and future directions for representations of functional brain organization. Nat. Neurosci. 23, 1484–1495 (2020).
Huertas, I. et al. A Bayesian spatial model for neuroimaging data based on biologically informed basis functions. Neuroimage 161, 134–148 (2017).
Kia, S. M. & Marquand, A. Normative modeling of neuroimaging data using scalable multi-task Gaussian processes. Preprint at arXiv https://doi.org/10.48550/arXiv.1806.01047 (2018).
Kia, S. M., Beckmann, C. F. & Marquand, A. F. Scalable multi-task Gaussian process tensor regression for normative modeling of structured variation in neuroimaging data. Preprint at arXiv https://doi.org/10.48550/arXiv.1808.00036 (2018).
Jahn, A. et al. Andy’s Brain Book https://andysbrainbook.readthedocs.io/en/latest (2020).
Casey, B. J. et al. The Adolescent Brain Cognitive Development (ABCD) study: imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 32, 43–54 (2018).
Thompson, P. M. et al. ENIGMA and global neuroscience: a decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl. Psychiatry 10, 100 (2020).
Beer, J. C. et al. Longitudinal ComBat: a method for harmonizing longitudinal multi-scanner imaging data. Neuroimage 220, 117129 (2020).
Fortin, J.-P. et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage 161, 149–170 (2017).
Fortin, J.-P. et al. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 167, 104–120 (2018).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Nygaard, V., Rødland, E. A. & Hovig, E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics 17, 29–39 (2016).
Noirhomme, Q. et al. Biased binomial assessment of cross-validated estimation of classification accuracies illustrated in diagnosis predictions. Neuroimage Clin. 4, 687–694 (2014).
Marquand, A. F., Wolfers, T., Mennes, M., Buitelaar, J. & Beckmann, C. F. Beyond lumping and splitting: a review of computational approaches for stratifying psychiatric disorders. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 1, 433–447 (2016).
Rahimi, A. & Recht, B. Random features for large-scale kernel machines. In NIPS'07: Proceedings of the 20th International Conference on Neural Information Processing Systems 1177–1184 (2007).
Lv, J. et al. Individual deviations from normative models of brain structure in a large cross-sectional schizophrenia cohort. Mol. Psychiatry 26, 3512–3523 (2021).
Snelson, E., Ghahramani, Z. & Rasmussen, C. Warped Gaussian processes. in Advances in Neural Information Processing Systems vol. 16 (MIT Press, 2004).
Hensman, J., Fusi, N. & Lawrence, N. D. Gaussian processes for big data. Preprint at arXiv https://doi.org/10.48550/arXiv.1309.6835 (2013).
Bethlehem, R. et al. Brain charts for the human lifespan. Nature https://doi.org/10.1038/s41586-022-04554-y (2022).
Rutherford, S. et al. Charting brain growth and aging at high spatial precision. eLife 11, e72904 (2022).
Rutherford, S. et al. The Normative Modeling Framework for Computational Psychiatry (Zenodo, 2021); https://doi.org/10.5281/zenodo.5592153
Acknowledgements
This research was supported by grants from the European Research Council (ERC, grant ‘MENTALPRECISION’ 10100118 and ‘BRAINMINT’ 802998), the Wellcome Trust under an Innovator award (‘BRAINCHART’, 215698/Z/19/Z) and a Strategic Award (098369/Z/12/Z), the Dutch Organisation for Scientific Research (VIDI grant 016.156.415). T.W. also gratefully acknowledges the Niels Stensen Fellowship as well as the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie Grant agreement no. 895011.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.R., S.M.K., T.W., C.F., M.Z., R.D., P.B., A.W., S.V., H.G.R., C.F.B. and A.F.M. Methodology: S.R., S.M.K., T.W., C.F., M.Z., R.D. and A.F.M. Data curation: S.R. and A.F.M. Writing—original draft: S.R. Writing—reviewing and editing: S.R., S.M.K., T.W., C.F., M.Z., R.D., P.B., A.W., S.V., H.G.R., C.F.B. and A.F.M. Visualization: S.R. Supervision: H.R., C.F.B. and A.F.M. Funding acquisition: H.R., C.F.B. and A.F.M.
Corresponding author
Ethics declarations
Competing interests
C.F.B. is director and shareholder of SBGNeuro Ltd. H.G.R. received speaker’s honorarium from Lundbeck and Janssen. The other authors report no conflicts of interest.
Peer review
Peer review information
Nature Protocols thanks Linden Parkes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Marquand, A. F. et al. Mol. Psychiatry 24, 1415–1424 (2019): https://doi.org/10.1038/s41380-019-0441-1
Zabihi, M. et al. Transl. Psychiatry 10, 384 (2020): https://doi.org/10.1038/s41398-020-01057-0
Wolfers, T. JAMA Psychiatry 75, 1146–1155 (2018): https://doi.org/10.1001/jamapsychiatry.2018.2467
Supplementary information
Rights and permissions
About this article
Cite this article
Rutherford, S., Kia, S.M., Wolfers, T. et al. The normative modeling framework for computational psychiatry. Nat Protoc 17, 1711–1734 (2022). https://doi.org/10.1038/s41596-022-00696-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00696-5
This article is cited by
-
Deep learning based joint fusion approach to exploit anatomical and functional brain information in autism spectrum disorders
Brain Informatics (2024)
-
Lifespan development of thalamic nuclei and characterizing thalamic nuclei abnormalities in schizophrenia using normative modeling
Neuropsychopharmacology (2024)
-
Population-wide cerebellar growth models of children and adolescents
Nature Communications (2024)
-
Population imaging cerebellar growth for personalized neuroscience
Nature Communications (2024)
-
Cortical thickness modeling and variability in Alzheimer’s disease and frontotemporal dementia
Journal of Neurology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.