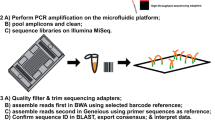
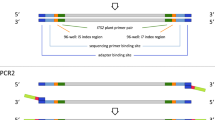
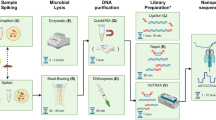
Abstract
In many parts of the world, human-mediated environmental change is depleting biodiversity faster than it can be characterized, while invasive species cause agricultural damage, threaten human health and disrupt native habitats. Consequently, the application of effective approaches for rapid surveillance and identification of biological specimens is increasingly important to inform conservation and biosurveillance efforts. Taxonomic assignments have been greatly advanced using sequence-based applications, such as DNA barcoding, a diagnostic technique that utilizes PCR and DNA sequence analysis of standardized genetic regions. However, in many biodiversity hotspots, endeavors are often hindered by a lack of laboratory infrastructure, funding for biodiversity research and restrictions on the transport of biological samples. A promising development is the advent of low-cost, miniaturized scientific equipment. Such tools can be assembled into functional laboratories to carry out genetic analyses in situ, at local institutions, field stations or classrooms. Here, we outline the steps required to perform amplicon sequencing applications, from DNA isolation to nanopore sequencing and downstream data analysis, all of which can be conducted outside of a conventional laboratory environment using miniaturized scientific equipment, without reliance on Internet connectivity. Depending on sample type, the protocol (from DNA extraction to full bioinformatic analyses) can be completed within 10 h, and with appropriate quality controls can be used for diagnostic identification of samples independent of core genomic facilities that are required for alternative methods.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Example data files can be found in the Supplementary Information.
Code availability
The code used can be found on the NGSpeciesID GitHub page: https://github.com/ksahlin/NGSpeciesID. The code in this protocol has been peer reviewed.
References
Dirzo, R. et al. Defaunation in the Anthropocene. Science 345, 401–406 (2014).
Barnosky, A. D. et al. Approaching a state shift in Earth’s biosphere. Nature 486, 52–58 (2012).
Diagne, C. et al. High and rising economic costs of biological invasions worldwide. Nature 592, 571–576 (2021).
Seebens, H. et al. Global rise in emerging alien species results from increased accessibility of new source pools. Proc. Natl Acad. Sci. USA 115, E2264–E2273 (2018).
Hebert, P. D. N., Ratnasingham, S. & de Waard, J. R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B 270, S96–S99 (2003).
Ratnasingham, S. & Hebert, P. bold: The Barcode of Life Data System. Mol. Ecol. Notes 7, 355–364 (2007).
Mizrachi, I. Chapter 1: GenBank: The Nucleotide Sequence Database (NCBI, 2013); https://www.ncbi.nlm.nih.gov/books/NBK470040/
Shokralla, S. et al. Massively parallel multiplex DNA sequencing for specimen identification using an Illumina MiSeq platform. Sci. Rep. 5, 9687 (2015).
Martinez, B. et al. Technology innovation: advancing capacities for the early detection of and rapid response to invasive species. Biol. Invasions 22, 75–100 (2020).
Krehenwinkel, H., Pomerantz, A. & Prost, S. Genetic biomonitoring and biodiversity assessment using portable sequencing technologies: current uses and future directions. Genes 10, 858 (2019).
Menegon, M. et al. On site DNA barcoding by nanopore sequencing. PLoS ONE 12, e0184741 (2017).
Pomerantz, A. et al. Real-time DNA barcoding in a rainforest using nanopore sequencing: opportunities for rapid biodiversity assessments and local capacity building. Gigascience 7, giy033 (2018).
Blanco, M. B. et al. Next-generation technologies applied to age-old challenges in Madagascar. Conserv. Genet. 21, 785–793 (2020).
Chang, J. J. M., Ip, Y. C. A., Ng, C. S. L. & Huang, D. Takeaways from mobile DNA barcoding with BentoLab and MinION. Genes 11, 1121 (2020).
Johnson, S. S., Zaikova, E., Goerlitz, D. S., Bai, Y. & Tighe, S. W. Real-time DNA sequencing in the Antarctic Dry Valleys using the Oxford Nanopore Sequencer. J. Biomol. Tech. 28, 2–7 (2017).
Quick, J. et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232 (2016).
Faria, N. R. et al. Mobile real-time surveillance of Zika virus in Brazil. Genome Med. 8, 97 (2016).
Watsa, M., Erkenswick, G. A., Pomerantz, A. & Prost, S. Portable sequencing as a teaching tool in conservation and biodiversity research. PLoS Biol. 18, e3000667 (2020).
Salazar, A. N. et al. An educational guide for nanopore sequencing in the classroom. PLoS Comput. Biol. 16, e1007314 (2020).
Jain, M., Olsen, H. E., Paten, B. & Akeson, M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 17, 239 (2016).
Weirather, J. L. et al. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Research 6, 1–32 (2017).
Wick, R. R., Judd, L. M. & Holt, K. E. Deepbinner: demultiplexing barcoded Oxford Nanopore reads with deep convolutional neural networks. PLoS Comput. Biol. 14, e1006583 (2018).
Krehenwinkel, H. et al. Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. Gigascience 8, giz006 (2019).
Srivathsan, A. et al. Rapid, large-scale species discovery in hyperdiverse taxa using 1D MinION sequencing. BMC Biol. 17, 96 (2019).
Vasiljevic, N. et al. Developmental validation of Oxford Nanopore Technology MinION sequence data and the NGSpeciesID bioinformatic pipeline for forensic genetic species identification. Forensic Sci. Int. Genet. 53, 102493 (2021).
Maestri, S. et al. A rapid and accurate MinION-based workflow for tracking species biodiversity in the field. Genes 10, 468 (2019).
Seah, A., Lim, M. C. W., McAloose, D., Prost, S. & Seimon, T. A. MinION-based DNA barcoding of preserved and non-invasively collected wildlife samples. Genes 11, 445 (2020).
Srivathsan, A. et al. MinION barcodes: biodiversity discovery and identification by everyone, for everyone. Preprint at BioRxiv https://doi.org/10.1101/2021.03.09.434692 (2021).
Atkins, P. A. P., Gamo, M. E. S. & Voytas, D. F. Analyzing plant gene targeting outcomes and conversion tracts with nanopore sequencing. Int. J. Mol. Sci. 22, 9723 (2021).
Simmons, D. R. et al. The Collection of Zoosporic Eufungi at the University of Michigan (CZEUM): introducing a new repository of barcoded Chytridiomyceta and Blastocladiomycota cultures. IMA Fungus 11, 20 (2020).
Sahlin, K., Lim, M. C. W. & Prost, S. NGSpeciesID: DNA barcode and amplicon consensus generation from long-read sequencing data. Ecol. Evol. 11, 1392–1398 (2021).
Taberlet, P. et al. Power and limitations of the chloroplast trn L (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14 (2007).
Dieffenbach, C. W., Lowe, T. M. & Dveksler, G. S. General concepts for PCR primer design. Genome Res. 3, S30–S37 (1993).
Singh, V. & Kumar, A. PCR primer design. Mol. Biol. Today 2, 27–32 (2001).
Untergasser, A. et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Ye, J. et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13, 134 (2012).
Bohmann, K. et al. Strategies for sample labelling and library preparation in DNA metabarcoding studies. Mol. Ecol. Res. 00, 1–16 (2021).
Smyth, R. P. et al. Reducing chimera formation during PCR amplification to ensure accurate genotyping. Gene 469, 45–51 (2010).
Zizka, V. M. A., Elbrecht, V., Macher, J.-N. & Leese, F. Assessing the influence of sample tagging and library preparation on DNA metabarcoding. Mol. Ecol. Resour. 19, 893–899 (2019).
Arulandhu, A. J. et al. Development and validation of a multi-locus DNA metabarcoding method to identify endangered species in complex samples. Gigascience 6, gix080 (2017).
Schnell, I. B., Bohmann, K. & Gilbert, M. T. P. Tag jumps illuminated—reducing sequence-to-sample misidentifications in metabarcoding studies. Mol. Ecol. Resour. 15, 1289–1303 (2015).
Lange, V. et al. Cost-efficient high-throughput HLA typing by MiSeq amplicon sequencing. BMC Genomics 15, 63 (2014).
Labrador, K., Agmata, A., Palermo, J. D., Follante, J. & Pante, Ma. J. Authentication of processed Philippine sardine products using Hotshot DNA extraction and minibarcode amplification. Food Control 98, 150–155 (2019).
Truett, G. E. et al. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and Tris (HotSHOT). BioTechniques 29, 52–54 (2000).
Knot, I. E., Zouganelis, G. D., Weedall, G. D., Wich, S. A. & Rae, R. DNA barcoding of nematodes using the MinION. Front. Ecol. Evol. https://doi.org/10.3389/fevo.2020.00100 (2020).
Wurzbacher, C. et al. Introducing ribosomal tandem repeat barcoding for fungi. Mol. Ecol. Resour. 19, 118–127 (2019).
Wilson, B. D., Eisenstein, M. & Soh, H. T. High-fidelity nanopore sequencing of ultra-short DNA targets. Anal. Chem. 91, 6783–6789 (2019).
Cornelis, S., Gansemans, Y., Deleye, L., Deforce, D. & Van Nieuwerburgh, F. Forensic SNP Genotyping using Nanopore MinION Sequencing. Sci. Rep. 7, 41759 (2017).
Srivathsan, A. et al. A MinIONTM-based pipeline for fast and cost-effective DNA barcoding. Mol. Ecol. Resour. 18, 1035–1049 (2018).
Sahlin, K. & Medvedev, P. in Research in Computational Molecular Biology (ed. Cowen, L. J.) 227–242 (Springer, 2019).
Vaser, R., Sović, I., Nagarajan, N. & Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017).
Daily, J. Parasail: SIMD C library for global, semi-global, and local pairwise sequence alignments. BMC Bioinform. 17, 81 (2016).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Byagathvalli, G., Pomerantz, A., Sinha, S., Standeven, J. & Bhamla, M. S. A 3D-printed hand-powered centrifuge for molecular biology. PLoS Biol. 17, e3000251 (2019).
Madden, T. Appendices. BLAST Command Line Applications User Manual [Internet] (Bethesda (MD): National Center for Biotechnology Information (US, 2021); https://www.ncbi.nlm.nih.gov/books/NBK279684/
Watsa, M., Wildlife Disease Surveillance Focus Group. Rigorous wildlife disease surveillance. Science 369, 145–147 (2020).
Acknowledgements
We thank L. Comai for the adjustment of the barcode_generator tool to work for our MinION indexing protocol. We thank ONT for providing technical support, and for making the offline MinKNOW software available to us. We also thank H. Asahara and the UC Berkeley DNA Sequencing Facility for technical support. We thank Tropical Herping, Rainforest Expeditions, Field Projects International and the Inkaterra Guides Field Station for support during fieldwork.
Author information
Authors and Affiliations
Contributions
A.P., N.V., A.S., E.H., S.K., H.K., S.W., R.O. and S.P. optimized and further developed the laboratory protocols. A.P. and S.P. tested and validated the approach and equipment in the field. K.S., M.L. and S.P. developed and validated the bioinformatics processing pipeline. R.O. and S.P. conceived the project. All authors wrote the manuscript and approved the contents of the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.P. became an employee of ONT PLC after the completion of the research described in the paper. The other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Simon Creer, Sujeevan Ratnasingham and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Pomerantz, A. et al. Gigascience 7, giy033 (2018): https://doi.org/10.1093/gigascience/giy033
Sahlin, K. et al. Ecol. Evol. 11, 1392–1398 (2021): https://doi.org/10.1002/ece3.7146
Vasiljevic, N. et al. Forensic Sci. Int. Genet. 53, 102493 (2021): https://doi.org/10.1016/j.fsigen.2021.102493
Supplementary information
Supplementary Data 1
An example MinION reads dataset generated using the outlined protocol (3,000 reads). The file contains reads of three fish species: the Atlantic cod (Gadus morhua), the Haddock (Melanogrammus aeglefinus) and the Whiting (Merlangius merlangus), sequenced on a Flongle flow cell. This is previously unpublished data.
Supplementary Data 2
An example index file for demultiplexing with minibar (can be used to demultiplex the example read data in Supplementary Data 1).
Supplementary Data 3
An example file of the primer sequences in fasta format (can be used for primer removal using NGSpeciesID).
Rights and permissions
About this article
Cite this article
Pomerantz, A., Sahlin, K., Vasiljevic, N. et al. Rapid in situ identification of biological specimens via DNA amplicon sequencing using miniaturized laboratory equipment. Nat Protoc 17, 1415–1443 (2022). https://doi.org/10.1038/s41596-022-00682-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00682-x
This article is cited by
-
Advances in environmental DNA monitoring: standardization, automation, and emerging technologies in aquatic ecosystems
Science China Life Sciences (2024)
-
Highly-multiplexed and efficient long-amplicon PacBio and Nanopore sequencing of hundreds of full mitochondrial genomes
BMC Genomics (2023)
-
Democratizing plant genomics to accelerate global food production
Nature Genetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.