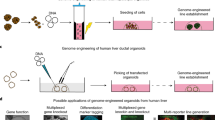
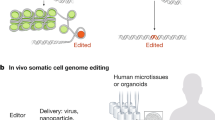
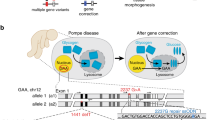
Abstract
Genetically engineered mouse models (GEMMs) transformed the study of organismal disease phenotypes but are limited by their lengthy generation in embryonic stem cells. Here, we describe methods for rapid and scalable genome engineering in somatic cells of the liver and pancreas through delivery of CRISPR components into living mice. We introduce the spectrum of genetic tools, delineate viral and nonviral CRISPR delivery strategies and describe a series of applications, ranging from gene editing and cancer modeling to chromosome engineering or CRISPR multiplexing and its spatio-temporal control. Beyond experimental design and execution, the protocol describes quantification of genetic and functional editing outcomes, including sequencing approaches, data analysis and interpretation. Compared to traditional knockout mice, somatic GEMMs face an increased risk for mouse-to-mouse variability because of the higher experimental demands of the procedures. The robust protocols described here will help unleash the full potential of somatic genome manipulation. Depending on the delivery method and envisaged application, the protocol takes 3–5 weeks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in either this paper or our original research study5,6. Source data are provided with this paper.
References
Bradley, A., Evans, M., Kaufman, M. H. & Robertson, E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255–256 (1984).
Doetschman, T. et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature 330, 576–578 (1987).
Kersten, K., de Visser, K. E., van Miltenburg, M. H. & Jonkers, J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 9, 137–153 (2017).
Weber, J. & Rad, R. Engineering CRISPR mouse models of cancer. Curr. Opin. Genet. Dev. 54, 88–96 (2019).
Weber, J. et al. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc. Natl Acad. Sci. USA 112, 13982–13987 (2015).
Maresch, R. et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat. Commun. 7, 10770 (2016).
Mueller, S. et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 554, 62–68 (2018).
Pickar-Oliver, A. & Gersbach, C. A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019).
Zhang, G., Budker, V. & Wolff, J. A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 10, 1735–1737 (1999).
Zhang, G. et al. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 11, 675–682 (2004).
Suda, T., Gao, X., Stolz, D. B. & Liu, D. Structural impact of hydrodynamic injection on mouse liver. Gene Ther. 14, 129–137 (2007).
Kim, M. J. & Ahituv, N. The hydrodynamic tail vein assay as a tool for the study of liver promoters and enhancers. Methods Mol. Biol. 1015, 279–289 (2013).
Liu, F., Song, Y. & Liu, D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6, 1258–1266 (1999).
Hubner, E. K. et al. Constitutive and inducible systems for genetic in vivo modification of mouse hepatocytes using hydrodynamic tail vein injection. J. Vis. Exp 2018, 56613 (2018).
Xue, W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014).
Sato, M. et al. Site-targeted non-viral gene delivery by direct DNA injection into the pancreatic parenchyma and subsequent in vivo electroporation in mice. Biotechnol. J. 8, 1355–1361 (2013).
Suzuki, T., Shin, B.-C., Fujikura, K., Matsuzaki, T. & Takata, K. Direct gene transfer into rat liver cells by in vivo electroporation. FEBS Lett. 425, 436–440 (1998).
Gürlevik, E. et al. Administration of gemcitabine after pancreatic tumor resection in mice induces an antitumor immune response mediated by natural killer cells. Gastroenterology 151, 338–350.e7 (2016).
Seehawer, M. et al. Necroptosis microenvironment directs lineage commitment in liver cancer. Nature 562, 69–75 (2018).
Rosenfeld, M. A. et al. Adenovirus-mediated transfer of a recombinant α1-antitrypsin gene to the lung epithelium in vivo. Science 252, 431–434 (1991).
Shirley, J. L., Jong, Y. P., de, Terhorst, C. & Herzog, R. W. Immune responses to viral gene therapy vectors. Mol. Ther. 28, 709–722 (2020).
Somanathan, S., Breous, E., Bell, P. & Wilson, J. M. AAV vectors avoid inflammatory signals necessary to render transduced hepatocyte targets for destructive T cells. Mol. Ther. 18, 977–982 (2010).
Snyder, R. O. et al. Correction of hemophilia B in canine and murine models using recombinant adeno-associated viral vectors. Nat. Med. 5, 64–70 (1999).
Cunningham, S. C., Dane, A. P., Spinoulas, A. & Alexander, I. E. Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol. Ther. 16, 1081–1088 (2008).
Fitzpatrick, Z. et al. Influence of pre-existing anti-capsid neutralizing and binding antibodies on AAV vector transduction. Mol. Ther. Methods Clin. Dev. 9, 119–129 (2018).
Grimm, D. et al. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood 102, 2412–2419 (2003).
Wu, Z., Asokan, A. & Samulski, R. J. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther. 14, 316–327 (2006).
Gao, G.-P. et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl Acad. Sci. USA 99, 11854–11859 (2002).
Inagaki, K. et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 14, 45–53 (2006).
Jimenez, V. et al. In vivo genetic engineering of murine pancreatic beta cells mediated by single-stranded adeno-associated viral vectors of serotypes 6, 8 and 9. Diabetologia 54, 1075–1086 (2011).
Quirin, K. A. et al. Safety and efficacy of AAV retrograde pancreatic ductal gene delivery in normal and pancreatic cancer mice. Mol. Ther. Methods Clin. Dev. 8, 8–20 (2018).
Penaud-Budloo, M. et al. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J. Virol. 82, 7875–7885 (2008).
Fisher, K. J. et al. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol. 70, 520–532 (1996).
Ferrari, F. K., Samulski, T., Shenk, T. & Samulski, R. J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70, 3227–3234 (1996).
Miao, C. H. et al. Nonrandom transduction of recombinant adeno-associated virus vectors in mouse hepatocytes in vivo: cell cycling does not influence hepatocyte transduction. J. Virol. 74, 3793–3803 (2000).
McCarty, D. M. et al. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 10, 2112–2118 (2003).
Wang, Z. et al. Widespread and stable pancreatic gene transfer by adeno-associated virus vectors via different routes. Diabetes 55, 875–884 (2006).
McCarty, D. M. Self-complementary AAV vectors; advances and applications. Mol. Ther. 16, 1648–1656 (2008).
Ideno, N. et al. A pipeline for rapidly generating genetically engineered mouse models of pancreatic cancer using in vivo CRISPR-Cas9-mediated somatic recombination. Lab. Invest. 99, 1233–1244 (2019).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Bell, P. et al. Inverse zonation of hepatocyte transduction with AAV vectors between mice and non-human primates. Mol. Genet. Metab. 104, 395–403 (2011).
Weber, J. et al. PiggyBac transposon tools for recessive screening identify B-cell lymphoma drivers in mice. Nat. Commun. 10, 1415 (2019).
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Chiou, S.-H. et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 29, 1576–1585 (2015).
Dow, L. E. et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 33, 390–394 (2015).
Bowling, S. et al. An engineered CRISPR-Cas9 mouse line for simultaneous readout of lineage histories and gene expression profiles in single cells. Cell 181, 1410–1422.e27 (2020).
Katigbak, A., Robert, F., Paquet, M. & Pelletier, J. Inducible genome editing with conditional CRISPR/Cas9 Mice. G3 (Bethesda) 8, 1627–1635 (2018).
Lundin, A. et al. Development of an ObLiGaRe Doxycycline Inducible Cas9 system for pre-clinical cancer drug discovery. Nat. Commun. 11, 4903 (2020).
Premsrirut, P. K. et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell 145, 145–158 (2011).
Bardeesy, N. et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl Acad. Sci. USA 103, 5947–5952 (2006).
Wang, G. et al. Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using AAV-CRISPR-mediated direct in vivo screening. Sci. Adv. 4, eaao5508 (2018).
Wei, T., Cheng, Q., Min, Y.-L., Olson, E. N. & Siegwart, D. J. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 11, 3232 (2020).
Blasco, R. B. et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 9, 1219–1227 (2014).
Maddalo, D. et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516, 423–427 (2014).
Rowley, M. et al. Inactivation of Brca2 promotes Trp53-associated but inhibits KrasG12D-dependent pancreatic cancer development in mice. Gastroenterology 140, 1303–1313.e1-3 (2011).
Xu, C. et al. piggyBac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc. Natl Acad. Sci. USA 114, 722–727 (2017).
Xu, Y., Liu, R. & Dai, Z. Key considerations in designing CRISPR/Cas9-carrying nanoparticles for therapeutic genome editing. Nanoscale 12, 21001–21014 (2020).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Mitchell, M. J. et al. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov 20, 101–124 (2020).
Sanità, G., Carrese, B. & Lamberti, A. Nanoparticle surface functionalization: how to improve biocompatibility and cellular internalization. Front. Mol. Biosci. 7, 587012 (2020).
Finn, J. D. et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22, 2227–2235 (2018).
Yin, H. et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 35, 1179–1187 (2017).
Zhang, Y.-N., Poon, W., Tavares, A. J., McGilvray, I. D. & Chan, W. C. W. Nanoparticle-liver interactions: cellular uptake and hepatobiliary elimination. J. Control. Release 240, 332–348 (2016).
Yang, Y. et al. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc. Natl Acad. Sci. USA 91, 4407–4411 (1994).
Worgall, S., Wolff, G., Falck-Pedersen, E. & Crystal, R. G. Innate immune mechanisms dominate elimination of adenoviral vectors following in vivo administration. Hum. Gene Ther. 8, 37–44 (1997).
Muruve, D. A. et al. Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo. J. Virol. 78, 5966–5972 (2004).
Alba, R., Bosch, A. & Chillon, M. Gutless adenovirus: last-generation adenovirus for gene therapy. Gene Ther. 12(Suppl 1), S18–S27 (2005).
Morró, M. et al. Pancreatic transduction by helper-dependent adenoviral vectors via intraductal delivery. Hum. Gene Ther. 25, 824–836 (2014).
Gallaher, S. D., Gil, J. S., Dorigo, O. & Berk, A. J. Robust in vivo transduction of a genetically stable Epstein-Barr virus episome to hepatocytes in mice by a hybrid viral vector. J. Virol. 83, 3249–3257 (2009).
Cronin, J., Zhang, X.-Y. & Reiser, J. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 5, 387–398 (2005).
Pan, D. et al. Biodistribution and toxicity studies of VSVG-pseudotyped lentiviral vector after intravenous administration in mice with the observation of in vivo transduction of bone marrow. Mol. Ther. 6, 19–29 (2002).
Pfeifer, A. et al. Transduction of liver cells by lentiviral vectors: analysis in living animals by fluorescence imaging. Mol. Ther. 3, 319–322 (2001).
van Til, N. P. et al. Kupffer cells and not liver sinusoidal endothelial cells prevent lentiviral transduction of hepatocytes. Mol. Ther. 11, 26–34 (2005).
Ranzani, M. et al. Lentiviral vector-based insertional mutagenesis identifies genes associated with liver cancer. Nat. Methods 10, 155–161 (2013).
Dalsgaard, T. et al. Improved lentiviral gene delivery to mouse liver by hydrodynamic vector injection through tail vein. Mol. Ther. Nucleic Acids 12, 672–683 (2018).
Brown, B. D. et al. In vivo administration of lentiviral vectors triggers a type I interferon response that restricts hepatocyte gene transfer and promotes vector clearance. Blood 109, 2797–2805 (2007).
Follenzi, A. et al. Targeting lentiviral vector expression to hepatocytes limits transgene-specific immune response and establishes long-term expression of human antihemophilic factor IX in mice. Blood 103, 3700–3709 (2004).
Annunziato, S. et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 30, 1470–1480 (2016).
Brown, B. D. et al. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood 110, 4144–4152 (2007).
Yin, H. et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 32, 551–553 (2014).
Winters, I. P. et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat. Commun. 8, 2053 (2017).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Kurt, I. C. et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 39, 41–46 (2020).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Chadwick, A. C., Wang, X. & Musunuru, K. In vivo base editing of PCSK9 (proprotein convertase subtilisin/kexin type 9) as a therapeutic alternative to genome editing. Arterioscler. Thromb. Vasc. Biol. 37, 1741–1747 (2017).
Villiger, L. et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 24, 1519–1525 (2018).
Liu, P. et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. 12, 2121 (2021).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Gemberling, M. P. et al. Transgenic mice for in vivo epigenome editing with CRISPR-based systems. Nat. Methods 18, 965–974 (2021).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Zhou, H. et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat. Neurosci. 21, 440–446 (2018).
Wangensteen, K. J. et al. Combinatorial genetics in liver repopulation and carcinogenesis with a in vivo CRISPR activation platform. Hepatology 68, 663–676 (2018).
Liao, H.-K. et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell 171, 1495–1507.e15 (2017).
Chew, W. L. et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 13, 868–874 (2016).
Moreno, A. M. et al. In situ gene therapy via AAV-CRISPR-Cas9-mediated targeted gene regulation. Mol. Ther. 26, 1818–1827 (2018).
Hilton, I. B. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015).
Liu, X. S. et al. Editing DNA methylation in the mammalian genome. Cell 167, 233–247.e17 (2016).
Cox, D. B. T. et al. RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017).
Abudayyeh, O. O. et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14 (2018).
Du, M., Jillette, N., Zhu, J. J., Li, S. & Cheng, A. W. CRISPR artificial splicing factors. Nat. Commun. 11, 2973 (2020).
Wilson, C., Chen, P. J., Miao, Z. & Liu, D. R. Programmable m6A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat. Biotechnol. 38, 1431–1440 (2020).
He, B. et al. Modulation of metabolic functions through Cas13d-mediated gene knockdown in liver. Protein Cell 11, 518–524 (2020).
Jiang, W. et al. Precise and efficient silencing of mutant KrasG12D by CRISPR-CasRx controls pancreatic cancer progression. Theranostics 10, 11507–11519 (2020).
Wang, D. et al. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum. Gene Ther. 26, 432–442 (2015).
Enache, O. M. et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 52, 662–668 (2020).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Akcakaya, P. et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416–419 (2018).
Chen, J. S. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Wong, N., Liu, W. & Wang, X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16, 218 (2015).
Yarrington, R. M., Verma, S., Schwartz, S., Trautman, J. K. & Carroll, D. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc. Natl Acad. Sci. USA 115, 9351–9358 (2018).
Hanna, R. E. & Doench, J. G. Design and analysis of CRISPR-Cas experiments. Nat. Biotechnol. 38, 813–823 (2020).
Montague, T. G., Cruz, J. M., Gagnon, J. A., Church, G. M. & Valen, E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407 (2014).
Heigwer, F., Kerr, G. & Boutros, M. E-CRISP: fast CRISPR target site identification. Nat. Methods 11, 122–123 (2014).
Concordet, J.-P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Meier, J. A., Zhang, F. & Sanjana, N. E. GUIDES: sgRNA design for loss-of-function screens. Nat. Methods 14, 831–832 (2017).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Park, J., Lim, K., Kim, J.-S. & Bae, S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33, 286–288 (2017).
Güell, M., Yang, L. & Church, G. M. Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA). Bioinformatics 30, 2968–2970 (2014).
Sledzinski, P., Nowaczyk, M. & Olejniczak, M. Computational tools and resources supporting CRISPR-Cas experiments. Cells 9, 1288 (2020).
Hess, J. et al. Gain of chromosome band 7q11 in papillary thyroid carcinomas of young patients is associated with exposure to low-dose irradiation. Proc. Natl Acad. Sci. USA 108, 9595–9600 (2011).
Lange, S. et al. Analysis pipelines for cancer genome sequencing in mice. Nat. Protoc. 15, 266–315 (2020).
Jentsch, I., Adler, I. D., Carter, N. P. & Speicher, M. R. Karyotyping mouse chromosomes by multiplex-FISH (M-FISH). Chromosome Res. 9, 211–214 (2001).
Espina, V. et al. Laser-capture microdissection. Nat. Protoc. 1, 586–603 (2006).
Bell, J. B. et al. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat. Protoc. 2, 3153–3165 (2007).
Sakai, M., Nishikawa, M., Thanaketpaisarn, O., Yamashita, F. & Hashida, M. Hepatocyte-targeted gene transfer by combination of vascularly delivered plasmid DNA and in vivo electroporation. Gene Ther. 12, 607–616 (2005).
Rizvi, F. et al. Murine liver repair via transient activation of regenerative pathways in hepatocytes using lipid nanoparticle-complexed nucleoside-modified mRNA. Nat. Commun. 12, 613 (2021).
Li, Q., Kay, M. A., Finegold, M., Stratford-Perricaudet, L. D. & Woo, S. L. Assessment of recombinant adenoviral vectors for hepatic gene therapy. Hum. Gene Ther. 4, 403–409 (1993).
Ryu, S.-M. et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 36, 536–539 (2018).
Thakore, P. I. et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat. Commun. 9, 1674 (2018).
Acknowledgements
D.S. is supported by the European Research Council (Consolidator Grant 648521) and the Deutsche Forschungsgemeinschaft (SA1374/4-2; SFB 1321 Project-ID 329628492, SFB 1371 Project-ID 395357507). R.R. is supported by the European Research Council (Consolidator Grants PACA-MET (819642) and MSCA-ITN-ETN (861196)), the Deutsche Forschungsgemeinschaft (DFG RA1629/2-1; SFB1321), the German Cancer Consortium and the Deutsche Krebshilfe (70114314).
Author information
Authors and Affiliations
Contributions
T.K., J.L., R.M., J.W., S.M., R.O. and R.R. conceptualized, designed or developed workflows, tools or procedures. U.E. provided resources and critical input to HTVI experiments. P.A., D.R., S. Brummer and S.E produced and purified AAV. T.K., J.L., R.M., J.W., S.M., R.O., N.G. and J.G. performed wet-lab experiments. N.A.K. performed computational analysis. A.A., J.M., M.S.-S., M.R., G.S. and D.S. provided biological resources. T.K, J.L. and R.R. wrote the manuscript with input from R.M., J.W., S.A.W., S. Bärthel, C.F., A.P. and C.J.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Takahiro Kodama and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Maresch, R. et al. Nat. Commun. 7, 10770 (2016): https://doi.org/10.1038/ncomms10770
Weber, J. et al. Proc. Natl Acad. Sci. USA 112, 13982–13987 (2015): https://doi.org/10.1073/pnas.1512392112
Mueller, S. et al. Nature 554, 62–68 (2018): https://doi.org/10.1038/nature25459
Supplementary information
Source data
Source Data Fig. 4c
Quantification of transduction efficiencies of scAAV8 in the pancreas and the liver in Rosa26mT/mG reporter mice.
Source Data Fig. 5b
Quantification of CRISPR editing efficiencies in the liver and pancreas upon scAAV8-based sgRNA delivery.
Source Data Fig. 6
Statistical data.
Rights and permissions
About this article
Cite this article
Kaltenbacher, T., Löprich, J., Maresch, R. et al. CRISPR somatic genome engineering and cancer modeling in the mouse pancreas and liver. Nat Protoc 17, 1142–1188 (2022). https://doi.org/10.1038/s41596-021-00677-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00677-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.