Abstract
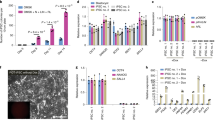
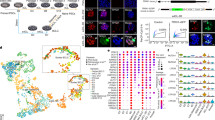
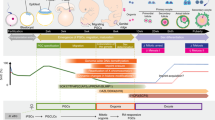
Naive human pluripotent stem cells (hPSCs) can be used to generate mature human cells of all three germ layers in mouse–human chimeric embryos. Here, we describe a protocol for generating mouse–human chimeric embryos by injecting naive hPSCs converted from the primed state. Primed hPSCs are treated with a mammalian target of rapamycin inhibitor (Torin1) for 3 h and dissociated to single cells, which are plated on mouse embryonic fibroblasts in 2iLI medium, a condition essentially the same for culturing mouse embryonic stem cells. After 3–4 d, bright, dome-shaped colonies with mouse embryonic stem cell morphology are passaged in 2iLI medium. Established naive hPSCs are injected into mouse blastocysts, which produce E17.5 mouse embryos containing 0.1–4.0% human cells as quantified by next-generation sequencing of 18S ribosomal DNA amplicons. The protocol is suitable for studying the development of hPSCs in mouse embryos and may facilitate the generation of human cells, tissues and organs in animals.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Tesar, P. J. et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 (2007).
Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Nichols, J. & Smith, A. Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 (2009).
Bradley, A., Evans, M., Kaufman, M. H. & Robertson, E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255–256 (1984).
James, D., Noggle, S. A., Swigut, T. & Brivanlou, A. H. Contribution of human embryonic stem cells to mouse blastocysts. Dev. Biol. 295, 90–102 (2006).
Bao, S. et al. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461, 1292–1295 (2009).
Murayama, H. et al. Successful reprogramming of epiblast stem cells by blocking nuclear localization of beta-catenin. Stem Cell Rep. 4, 103–113 (2015).
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Gafni, O. et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286 (2013).
Chan, Y. S. et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13, 663–675 (2013).
Theunissen, T. W. et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 524–526 (2014).
Ware, C. B. et al. Derivation of naive human embryonic stem cells. Proc. Natl Acad. Sci. USA 111, 4484–4489 (2014).
Wu, J. et al. Interspecies chimerism with mammalian pluripotent stem cells. Cell 168, 473–486 e415 (2017).
Yang, Y. et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 169, 243–257 e225 (2017).
Weinberger, L., Ayyash, M., Novershtern, N. & Hanna, J. H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 17, 155–169 (2016).
Wang, Y. & Gao, S. Human naive embryonic stem cells: how full is the glass? Cell Stem Cell 18, 301–303 (2016).
Hu, Z. et al. Transient inhibition of mTOR in human pluripotent stem cells enables robust formation of mouse-human chimeric embryos. Sci. Adv. 6, eaaz0298 (2020).
Buecker, C. et al. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell 6, 535–546 (2010).
Hanna, J. et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl Acad. Sci. USA 107, 9222–9227 (2010).
Wang, W. et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl Acad. Sci. USA 108, 18283–18288 (2011).
Hu, Z. et al. Generation of naivetropic induced pluripotent stem cells from Parkinson’s disease patients for high-efficiency genetic manipulation and disease modeling. Stem Cells Dev. 24, 2591–2604 (2015).
Takashima, Y. et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269 (2014).
Huang, K., Maruyama, T. & Fan, G. The naive state of human pluripotent stem cells: a synthesis of stem cell and preimplantation embryo transcriptome analyses. Cell Stem Cell 15, 410–415 (2014).
Theunissen, T. W. et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19, 502–515 (2016).
Yang, J. et al. Establishment of mouse expanded potential stem cells. Nature 550, 393–397 (2017).
Gao, X. et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 21, 687–699 (2019).
Betschinger, J. et al. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153, 335–347 (2013).
Raben, N. & Puertollano, R. TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu. Rev. Cell Dev. Biol (2016).
Martina, J. A. et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal 7, ra9 (2014).
Cohen, M. A. et al. Human neural crest cells contribute to coat pigmentation in interspecies chimeras after in utero injection into mouse embryos. Proc. Natl Acad. Sci. USA 113, 1570–1575 (2016).
Stults, D. M., Killen, M. W., Pierce, H. H. & Pierce, A. J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 18, 13–18 (2008).
Gonzalez, I. L. & Schmickel, R. D. The human 18S ribosomal RNA gene: evolution and stability. Am. J. Hum. Genet 38, 419–427 (1986).
Kobayashi, T. et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142, 787–799 (2010).
Yamaguchi, T. et al. Interspecies organogenesis generates autologous functional islets. Nature 542, 191–196 (2017).
Wu, J. et al. Stem cells and interspecies chimaeras. Nature 540, 51–59 (2016).
Villegas, F. et al. Lysosomal signaling licenses embryonic stem cell differentiation via inactivation of Tfe3. Cell Stem Cell 24, 257–270 (2019).
Gibbons, J. G., Branco, A. T., Godinho, S. A., Yu, S. & Lemos, B. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc. Natl Acad. Sci. USA 112, 2485–2490 (2015).
Waites, K. B., Xiao, L., Liu, Y., Balish, M. F. & Atkinson, T. P. Mycoplasma pneumoniae from the respiratory tract and beyond. Clin. Microbiol. Rev. 30, 747–809 (2017).
Nagy, A. Manipulating the Mouse Embryo: A Laboratory Manual 3rd edn (Cold Spring Harbor Laboratory Press, 2003).
Uphoff, C. C. & Drexler, H. G. Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol. Biol. 731, 93–103 (2011).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahe, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Czechanski, A. et al. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc. 9, 559–574 (2014).
Okita, K. et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412 (2011).
Okita, K., Hong, H., Takahashi, K. & Yamanaka, S. Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat. Protoc. 5, 418–428 (2010).
Acknowledgements
We thank D. Barnas at the Gene Targeting and Transgenic Resource of Roswell Park Comprehensive Cancer Center for injections and transfers of mouse embryos. The work was supported by NYSTEM contracts C028129 (J.F.), C029556 (J.F.) and C30290GG (fellowship for H.L.) and the Buffalo Blue Sky Initiative (J.F.). We thank E.C. Fisher and M.A. Juncker for reading the manuscript.
Author information
Authors and Affiliations
Contributions
B.Z. and H.L. contributed equally to the study. B.Z. developed the conversion protocol based on the initial method of Z.H. H.L. developed the method to analyze chimeric embryos by NGS. H.J. contributed to the analyses of naive hPSCs and chimeric embryos. A.B.S. performed blastocyst injections and embryo transfers. B.J.M. and D.A.Y. performed bioinformatics analysis of NGS data. J.F. conceived and supervised the study. All authors contributed to writing the paper.
Corresponding author
Ethics declarations
Competing interests
J.F. is a cofounder of Vitropy, LLC and ASDDR, LLC, and has a patent application regarding this work (no. 16/346534). All other authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Jacob Hanna and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Hu, Z. et al. Sci Adv 6, eaaz0298 (2020): https://doi.org/10.1126/sciadv.aaz0298
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, B., Li, H., Hu, Z. et al. Generation of mouse–human chimeric embryos. Nat Protoc 16, 3954–3980 (2021). https://doi.org/10.1038/s41596-021-00565-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00565-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.