Abstract
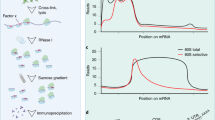
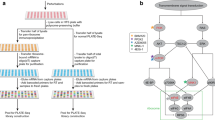
Several essential components of the electron transport chain, the major producer of ATP in mammalian cells, are encoded in the mitochondrial genome. These 13 proteins are translated within mitochondria by ‘mitoribosomes’. Defective mitochondrial translation underlies multiple inborn errors of metabolism and has been implicated in pathologies such as aging, metabolic syndrome and cancer. Here, we provide a detailed ribosome profiling protocol optimized to interrogate mitochondrial translation in mammalian cells (MitoRiboSeq), wherein mitoribosome footprints are generated with micrococcal nuclease and mitoribosomes are separated from cytosolic ribosomes and other RNAs by ultracentrifugation in a single straightforward step. We highlight critical steps during library preparation and provide a step-by-step guide to data analysis accompanied by open-source bioinformatic code. Our method outputs mitoribosome footprints at single-codon resolution. Codons with high footprint densities are sites of mitoribosome stalling. We recently applied this approach to demonstrate that defects in mitochondrial serine catabolism or in mitochondrial tRNA methylation cause stalling of mitoribosomes at specific codons. Our method can be applied to study basic mitochondrial biology or to characterize abnormalities in mitochondrial translation in patients with mitochondrial disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The raw FASTQ data files are deposited at NCBI’s Sequence Read Archive (SRA). The sequencing results used in the bioinformatics analysis are available under BioProject PRJNA590503 and SRA accession numbers SRR10491343, SRR10491342, SRR10491341 and SRR10491340.
Code availability
All related codes required for making the plots shown in this protocol are provided on the GitHub page at https://github.com/sophiahjli/MitoRiboSeq. These files are sufficient for readers to reproduce the bioinformatics results shown in this manuscript. The user can use the readme file with a step-by-step guide to set up the computation environment and to run the codes.
References
Spinelli, J. B. & Haigis, M. C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 20, 745–754 (2018).
Fernández-Vizarra, E., Tiranti, V. & Zeviani, M. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta 1793, 200–211 (2009).
Fox, T. D. Mitochondrial protein synthesis, import, and assembly. Genetics 192, 1203–1234 (2012).
Calvo, S. E. & Mootha, V. K. The mitochondrial proteome and human disease. Annu. Rev. Genom. Hum. G. 11, 25–44 (2010).
Greber, B. J. & Ban, N. Structure and function of the mitochondrial ribosome. Annu. Rev. Biochem. 85, 1–30 (2015).
DiMauro, S. & Schon, E. A. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348, 2656–2668 (2003).
Boczonadi, V. & Horvath, R. Mitochondria: impaired mitochondrial translation in human disease. Int. J. Biochem. Cell Biol. 48, 77–84 (2014).
Rötig, A. Human diseases with impaired mitochondrial protein synthesis. Biochim. Biophys. Acta 1807, 1198–1205 (2011).
Keilland, E. et al. The expanding phenotype of MELAS caused by the m.3291T > C mutation in the MT-TL1 gene. Mol. Genet. Metab. Rep. 6, 64–69 (2016).
Kirino, Y., Goto, Y., Campos, Y., Arenas, J. & Suzuki, T. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc. Natl Acad. Sci. USA 102, 7127–7132 (2005).
Škrtić, M. et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 20, 674–688 (2011).
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Wallace, D. C. Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698 (2012).
Dykens, J. A. & Will, Y. The significance of mitochondrial toxicity testing in drug development. Drug Discov. Today 12, 777–785 (2007).
Kalghatgi, S. et al. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in mammalian cells. Sci. Transl. Med. 5, 192ra85 (2013).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Ji, Z., Song, R., Regev, A. & Struhl, K. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4, e08890 (2015).
Chen, J. et al. Pervasive functional translation of noncanonical human open reading frames. Science 367, 1140–1146 (2020).
Jan, C. H., Williams, C. C. & Weissman, J. S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346, 1257521 (2014).
Williams, C. C., Jan, C. H. & Weissman, J. S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346, 748–751 (2014).
Loayza-Puch, F. et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature 530, 490–494 (2016).
Li, G.-W., Burkhardt, D., Gross, C. & Weissman, J. S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014).
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011).
Juntawong, P., Girke, T., Bazin, J. & Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl Acad. Sci. USA 111, E203–E212 (2014).
Itzhak, D. N., Tyanova, S., Cox, J. & Borner, G. H. Global, quantitative and dynamic mapping of protein subcellular localization. eLife 5, e16950 (2016).
Wolf, S. F. & Schlessinger, D. Nuclear metabolism of ribosomal RNA in growing, methionine-limited, and ethionine-treated HeLa cells. Biochemistry 16, 2783–2791 (1977).
Amunts, A., Brown, A., Toots, J., Scheres, S. H. W. & Ramakrishnan, V. The structure of the human mitochondrial ribosome. Science 348, 95–98 (2015).
Kummer, E. et al. Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 560, 263–267 (2018).
Montoya, J., Ojala, D. & Attardi, G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature 290, 290465a0 (1981).
Temperley, R. J., Wydro, M., Lightowlers, R. N. & Chrzanowska-Lightowlers, Z. M. Human mitochondrial mRNAs—like members of all families, similar but different. Biochim. Biophys. Acta 1797, 1081–1085 (2010).
Morscher, R. J. et al. Mitochondrial translation requires folate-dependent tRNA methylation. Nature 554, 128 (2018).
Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M. & Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012).
McGlincy, N. J. & Ingolia, N. T. Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129 (2017).
Johnson, G. E. & Li, G.-W. Genome-wide quantitation of protein synthesis rates in bacteria. Methods Enzymol. 612, 225–249 (2018).
Rooijers, K., Loayza-Puch, F., Nijtmans, L. G. & Agami, R. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat. Commun. 4, 2886 (2013).
Couvillion, M. T. & Churchman, L. S. Mitochondrial ribosome (mitoribosome) profiling for monitoring mitochondrial translation in vivo. Curr. Protoc. Mol. Biol. 119, 4.28.1–4.28.25 (2017).
Couvillion, M. T., Soto, I. C., Shipkovenska, G. & Churchman, L. S. Synchronized mitochondrial and cytosolic translation programs. Nature 533, 499 (2016).
Pearce, S. F. et al. Maturation of selected human mitochondrial tRNAs requires deadenylation. eLife 6, e27596 (2017).
Santos, D. A., Shi, L., Tu, B. P. & Weissman, J. S. Cycloheximide can distort measurements of mRNA levels and translation efficiency. Nucleic Acids Res. 47, 4974–4985 (2019).
Jones, C. N., Miller, C., Tenenbaum, A., Spremulli, L. L. & Saada, A. Antibiotic effects on mitochondrial translation and in patients with mitochondrial translational defects. Mitochondrion 9, 429–437 (2009).
Datta, A. K. & Burma, D. P. Association of ribonuclease I with ribosomes and their subunits. J Biol. Chem. 247, 6795–6801 (1972).
Busch, J. D. et al. MitoRibo-Tag mice provide a tool for in vivo studies of mitoribosome composition. Cell Rep. 29, 1728–1738.e9 (2019).
Culviner, P. H., Guegler, C. K. & Laub, M. T. A simple, cost-effective, and robust method for rRNA depletion in RNA-sequencing studies. mBio 11, e00010-20 (2020).
Sendoel, A. et al. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541, 494–499 (2017).
Riba, A. et al. Protein synthesis rates and ribosome occupancies reveal determinants of translation elongation rates. Proc. Natl Acad. Sci. USA 116, 15023–15032 (2019).
Mohammad, F., Green, R. & Buskirk, A. R. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. eLife 8, e42591 (2019).
Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018).
Michel, A. M. et al. RiboGalaxy: a browser based platform for the alignment, analysis and visualization of ribosome profiling data. RNA Biol. 13, 316–319 (2016).
Köster, J. & Rahmann, S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 34, 3600–3600 (2018).
Grüning, B. et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods 15, 475–476 (2018).
Dunn, J. G. & Weissman, J. S. Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC Genomics 17, 958 (2016).
Hwang, J.-Y. & Buskirk, A. R. A ribosome profiling study of mRNA cleavage by the endonuclease RelE. Nucleic Acids Res. 45, 327–336 (2017).
Woolstenhulme, C. J., Guydosh, N. R., Green, R. & Buskirk, A. R. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 11, 13–21 (2015).
Oh, E. et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308 (2011).
Taanman, J.-W. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta 1410, 103–123 (1999).
D’Souza, A. R. & Minczuk, M. Mitochondrial transcription and translation: overview. Essays Biochem. 62, 309–320 (2018).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Yates, A. D. et al. Ensembl 2020. Nucleic Acids Res. 48, D682–D688 (2019).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
R Core Team. R: a language and environment for statistical computing. https://www.r-project.org
Wickham, H. et al. Welcome to the tidyverse. J. Open Source Softw. 4, 1686 (2019).
Kolde, R. pheatmap: pretty heatmaps. https://rdrr.io/cran/pheatmap/
Slowikowski, K. ggrepel: automatically position non-overlapping text labels with “ggplot2” (2020).
Wilke, C. O. cowplot: streamlined plot theme and plot annotations for “ggplot2” (2019).
Gao, F. et al. Using mitoribosomal profiling to investigate human mitochondrial translation. Wellcome Open Res. 2, 116 (2018).
Acknowledgements
We thank R. J. Morscher for his help with establishing the initial MitoRiboSeq method. We thank the Gitai lab and the Rabinowitz lab for their comments and suggestions. We thank S. Vianello for his support with the artistic illustration of the protocol workflow. This work is supported by funding to J.D.R. from the US National Institutes of Health (NIH) (R01CA163591 and DP1DK113643) and StandUp to Cancer (SU2CAACR-DT-20-16). Z.G. was supported by the NIH (DP1AI124669).
Author information
Authors and Affiliations
Contributions
S.H.-J.L. and M.N. developed the protocol. S.H.-J.L. and L.P. developed the bioinformatics analysis pipeline. S.H.-J.L., M.N. and Z.G. wrote the paper with help from J.D.R.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Jean-Michel Cioni, Norbert Huebner and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Morscher, R. et al. Nature 554, 128–132 (2018): https://doi.org/10.1038/nature25460
Rights and permissions
About this article
Cite this article
Li, S.HJ., Nofal, M., Parsons, L.R. et al. Monitoring mammalian mitochondrial translation with MitoRiboSeq. Nat Protoc 16, 2802–2825 (2021). https://doi.org/10.1038/s41596-021-00517-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00517-1
This article is cited by
-
Human mitochondria require mtRF1 for translation termination at non-canonical stop codons
Nature Communications (2023)
-
Principles, challenges, and advances in ribosome profiling: from bulk to low-input and single-cell analysis
Advanced Biotechnology (2023)
-
A custom library construction method for super-resolution ribosome profiling in Arabidopsis
Plant Methods (2022)
-
Balanced mitochondrial and cytosolic translatomes underlie the biogenesis of human respiratory complexes
Genome Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.