Abstract
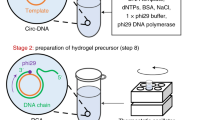
Circulating tumor cells (CTCs) enable noninvasive liquid biopsy and identification of cancer. Various approaches exist for the capture and release of CTCs, including microfluidic methods and those involving magnetic beads or nanostructured solid interfaces. However, the concomitant cell damage and fragmentation that often occur during capture make it difficult to extensively characterize and analyze living CTCs. Here, we describe an aptamer-trigger-clamped hybridization chain reaction (atcHCR) method for the capture of CTCs by porous 3D DNA hydrogels. The 3D environment of the DNA networks minimizes cell damage, and the CTCs can subsequently be released for live-cell analysis. In this protocol, initiator DNAs with aptamer-toehold biblocks specifically bind to the epithelial cell adhesion molecule (EpCAM) on the surface of CTCs, which triggers the atcHCR and the formation of a DNA hydrogel. The DNA hydrogel cloaks the CTCs, facilitating quantification with minimal cell damage. This method can be used to quantitively identify as few as 10 MCF-7 cells in a 2-µL blood sample. Decloaking of tumor cells via gentle chemical stimulus (ATP) is used to release living tumor cells for subsequent cell culture and live-cell analysis. We also describe how to use the protocol to encapsulate and release cells of cancer cell lines, which can be used in preliminary experiments to model CTCs. The whole protocol takes ~2.5 d to complete, including downstream cell culture and analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The main data supporting the examples of this protocol are available within the article and its Supplementary Information files. Extra data are available from the corresponding author upon reasonable request. The source data underlying Figs. 3e,f, 6d,e and 7c–f and Supplementary Figs. 2, 3, 5, 6b, 8a–c and 9 are provided as source data files.
References
Nagrath, S. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450, 1235–1239 (2007).
Hou, S. et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv. Mater. 25, 1547–1551 (2013).
Ke, Z. F. et al. Programming thermoresponsiveness of nano velcro substrates enables effective purification of circulating tumor cells in lung cancer patients. ACS Nano 9, 62–70 (2015).
Ozkumur, E. et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 5, 179ra147 (2013).
Stott, S. L. et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl Acad. Sci. USA 107, 18392–18397 (2010).
Zhou, G. B. et al. Multivalent capture and detection of cancer cells with DNA nanostructured biosensors and multibranched hybridization chain reaction amplification. Anal. Chem. 86, 7843–7848 (2014).
Yoon, H. J. et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 8, 735–741 (2013).
Zhang, P. C. et al. Programmable fractal nanostructured interfaces for specific recognition and electrochemical release of cancer cells. Adv. Mater. 25, 3566–3570 (2013).
Zhao, W. A. et al. Bioinspired multivalent DNA network for capture and release of cells. Proc. Natl Acad. Sci. USA 109, 19626–19631 (2012).
Cushing, M. C. & Anseth, K. S. Materials science. Hydrogel cell cultures. Science 316, 1133–1134 (2007).
Li, J. & Mooney, D. J. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 1, 16071 (2016).
Li, J. et al. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J. Am. Chem. Soc. 137, 1412–1415 (2015).
Seliktar, D. Designing cell-compatible hydrogels for biomedical applications. Science 336, 1124–1128 (2012).
Shen, Q. L. et al. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv. Mater. 25, 2368–2373 (2013).
Liu, Q. et al. Valency-controlled framework nucleic acid signal amplifiers. Angew. Chem. Int. Ed. 57, 7131–7135 (2018).
Guo, W. W. et al. pH-stimulated DNA hydrogels exhibiting shape-memory properties. Adv. Mater. 27, 73–78 (2015).
Guo, W. W. et al. Switchable bifunctional stimuli-triggered poly-N-isopropylacrylamide/DNA hydrogels. Angew. Chem. Inter. Ed. 53, 10134–10138 (2014).
Jin, J. et al. A triggered DNA hydrogel cover to envelop and release single cells. Adv. Mater. 25, 4714–4717 (2013).
Xing, Y. Z. et al. Self-assembled DNA hydrogels with designable thermal and enzymatic responsiveness. Adv. Mater. 23, 1117–1121 (2011).
Zhu, Z. et al. Au@Pt nanoparticle encapsulated target-responsive hydrogel with volumetric bar-chart chip readout for quantitative point-of-care testing. Angew. Chem. Int. Ed. 53, 12503–12507 (2014).
Song, Y. L. et al. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 85, 4141–4149 (2013).
Song, P. et al. DNA hydrogel with aptamer-toehold-based recognition, cloaking, and decloaking of circulating tumor cells for live cell analysis. Nano Lett. 17, 5193–5198 (2017).
Chen, X. Q. et al. Ultrasensitive electrochemical detection of prostate-specific antigen by using antibodies anchored on a DNA nanostructural scaffold. Anal. Chem. 86, 7337–7342 (2014).
Ge, Z. L. et al. Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor. Anal. Chem. 86, 2124–2130 (2014).
Ge, Z. L., Pei, H., Wang, L. H., Song, S. P. & Fan, C. H. Electrochemical single nucleotide polymorphisms genotyping on surface immobilized three-dimensional branched DNA nanostructure. Sci. China Chem. 54, 1273–1276 (2011).
Lin, M. H. et al. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. Int. Ed. 54, 2151–2155 (2015).
Lin, M. H. et al. Target-responsive, DNA nanostructure-based e-DNA sensor for microRNA analysis. Anal. Chem. 86, 2285–2288 (2014).
Pei, H. et al. A DNA Nanostructure-based biomolecular probe carrier platform for electrochemical biosensing. Adv. Mater. 22, 4754–4758 (2010).
Shen, J. et al. Valence-engineering of quantum dots using programmable DNA scaffolds. Angew. Chem. Int. Ed. 56, 16077–16081 (2017).
Ye, D. K., Zuo, X. L. & Fan, C. H. DNA nanostructure-based engineering of the biosensing interface for biomolecular detection. Prog. Chem. 29, 36–46 (2017).
Zhu, D. et al. A surface-confined proton-driven DNA pump using a dynamic 3D DNA scaffold. Adv. Mater. 28, 6860–6865 (2016).
Dirks, R. M. & Pierce, N. A. Triggered amplification by hybridization chain reaction. Proc. Natl Acad. Sci. USA 101, 15275–15278 (2004).
Wang, J. et al. Clamped hybridization chain reactions for the self-assembly of patterned DNA hydrogels. Angew. Chem. Int. Ed. 56, 2171–2175 (2017).
Li, C. et al. Rapid formation of a supramolecular polypeptide-DNA hydrogel for in situ three-dimensional multilayer bioprinting. Angew. Chem. Int. Ed. 54, 3957–3961 (2015).
Zhu, Z. et al. An aptamer cross-linked hydrogel as a colorimetric platform for visual detection. Angew. Chem. Int. Ed. 49, 1052–1056 (2010).
Kosuri, S. & Church, G. M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods 11, 499–507 (2014).
Acknowledgements
This work was financially supported by the Ministry of Science and Technology of China (2016YFA0201200), the National Natural Science Foundation of China (21904086, 21804088, 21804091), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20171913), the “Shuguang Program” supported by the Shanghai Education Development Foundation, and the Shanghai Municipal Education Commission (18SG16).
Author information
Authors and Affiliations
Contributions
X. Zuo, Q.L. and C.F. supervised the projects; D.Y., M.L., T.Z., P.S., L.S., H.W., X.M., X. Zuo and C.F. designed and conducted the experiments; F.W., X. Zhang, J.S., Z.G., L.W. and Q.L. analyzed the data; and Q.L., X. Zuo and C.F. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Song, P. et al. Nano Lett. 17, 5193–5198 (2017): https://doi.org/10.1021/acs.nanolett.7b01006
Key data used in this protocol
Song, P. et al. Nano Lett. 17, 5193–5198 (2017): https://doi.org/10.1021/acs.nanolett.7b01006
Supplementary information
Supplementary Information
Supplementary Figures 1–9.
Supplementary Video 1
E. coli movement in PBS
Supplementary Video 2
E. coli movement in DNA hydrogel
Supplementary Data 1
Statistical source data for Supplementary Figure 2
Supplementary Data 2
Statistical source data for Supplementary Figure 3
Supplementary Data 3
Statistical source data for Supplementary Figure 5
Supplementary Data 4
Statistical source data for Supplementary Figure 6
Supplementary Data 5
Statistical source data for Supplementary Figure 8
Supplementary Data 6
Statistical source data for Supplementary Figure 9
Source data
Source Data Fig. 3
Statistical source data
Source Data Fig. 4
Unprocessed gels
Source Data Fig. 6
Statistical source data
Source Data Fig. 7
Statistical source data
Rights and permissions
About this article
Cite this article
Ye, D., Li, M., Zhai, T. et al. Encapsulation and release of living tumor cells using hydrogels with the hybridization chain reaction. Nat Protoc 15, 2163–2185 (2020). https://doi.org/10.1038/s41596-020-0326-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0326-4
This article is cited by
-
Recent advances in photoelectrochemistry-coupled dual-modal biosensors: From constructions to biosensing applications
Nano Research (2024)
-
A temporally resolved DNA framework state machine in living cells
Nature Machine Intelligence (2023)
-
Current hydrogel advances in physicochemical and biological response-driven biomedical application diversity
Signal Transduction and Targeted Therapy (2021)
-
Rolling circle amplification (RCA)-based DNA hydrogel
Nature Protocols (2021)
-
Encapsulation and Release of Circulating Tumor Cells Using 3D DNA Hydrogels
Chemical Research in Chinese Universities (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.