Abstract
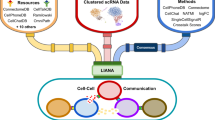
Cell–cell communication mediated by ligand–receptor complexes is critical to coordinating diverse biological processes, such as development, differentiation and inflammation. To investigate how the context-dependent crosstalk of different cell types enables physiological processes to proceed, we developed CellPhoneDB, a novel repository of ligands, receptors and their interactions. In contrast to other repositories, our database takes into account the subunit architecture of both ligands and receptors, representing heteromeric complexes accurately. We integrated our resource with a statistical framework that predicts enriched cellular interactions between two cell types from single-cell transcriptomics data. Here, we outline the structure and content of our repository, provide procedures for inferring cell–cell communication networks from single-cell RNA sequencing data and present a practical step-by-step guide to help implement the protocol. CellPhoneDB v.2.0 is an updated version of our resource that incorporates additional functionalities to enable users to introduce new interacting molecules and reduces the time and resources needed to interrogate large datasets. CellPhoneDB v.2.0 is publicly available, both as code and as a user-friendly web interface; it can be used by both experts and researchers with little experience in computational genomics. In our protocol, we demonstrate how to evaluate meaningful biological interactions with CellPhoneDB v.2.0 using published datasets. This protocol typically takes ~2 h to complete, from installation to statistical analysis and visualization, for a dataset of ~10 GB, 10,000 cells and 19 cell types, and using five threads.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The decidua and placenta datasets can be downloaded from ArrayExpress, with experiment code E-MTAB-6701.
Code availability
The CellPhoneDB code is available at https://github.com/Teichlab/cellphonedb. It can also be downloaded from https://cellphonedb.org/downloads. The code in this paper has been peer-reviewed.
References
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Ramilowski, J. A. et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 6, 7866 (2015).
Hie, B., Cho, H., DeMeo, B., Bryson, B. & Berger, B. Geometric sketching compactly summarizes the single-cell transcriptomic landscape. Cell Syst. 8, 483–493.e7 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018).
Braga, F. A. V. et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 25, 1153–1163 (2019).
Stewart, B. J. et al. Spatiotemporal immune zonation of the human kidney. Science 365, 1461–1466 (2019).
Popescu, D.-M. et al. Decoding the development of the blood and immune systems during human fetal liver haematopoiesis. Nature 574, 365–371 (2019).
Davidson, S. et al. Single-cell RNA sequencing reveals a dynamic stromal niche within the evolving tumour microenvironment. Preprint at bioRxiv: https://doi.org/10.1101/467225 (2018)
Skelly, D. A. et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 22, 600–610 (2018).
Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538 (2017).
Pavličev, M. et al. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res. 27, 349–361 (2017).
Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e24 (2017).
Suryawanshi, H. et al. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4, eaau4788 (2018).
Zhou, J. X., Taramelli, R., Pedrini, E., Knijnenburg, T. & Huang, S. Author correction: Extracting intercellular signaling network of cancer tissues using ligand-receptor expression patterns from whole-tumor and single-cell transcriptomes. Sci. Rep. 8, 17903 (2018).
Cohen, M. et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell 175, 1031–1044.e18 (2018).
Kumar, M. P. et al. Analysis of single-cell rna-seq identifies cell-cell communication associated with tumor characteristics. Cell Rep. 25, 1458–1468.e4 (2018).
Halpern, K. B. et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 36, 962–970 (2018).
Joost, S. et al. Single-cell transcriptomics of traced epidermal and hair follicle stem cells reveals rapid adaptations during wound healing. Cell Rep. 25, 585–597.e7 (2018).
Boisset, J.-C. et al. Mapping the physical network of cellular interactions. Nat. Methods 15, 547–553 (2018).
Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M. & Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Svensson, V. A method for transcriptome-wide gene expression quantification in intact tissues. Immunol. Cell Biol. 97, 439–441 (2019).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Proteomics Standards Initiative. Proteomics Standards Initiative common query interface in Encyclopedia of Systems Biology (eds Dubitzky, W., Wolkenhauer, O., Cho, K.H. & Yokota, H.) 1798–1798 (Springer, 2013): https://doi.org/10.1007/978-1-4419-9863-7_101243
Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods 9, 345–350 (2012).
Orchard, S. et al. The MIntAct project-IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363 (2014).
Breuer, K. et al. InnateDB: systems biology of innate immunity and beyond-recent updates and continuing curation. Nucleic Acids Res. 41, D1228–D1233 (2013).
Clerc, O. et al. MatrixDB: integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 47, D376–D381 (2019).
Licata, L. et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 40, D857–D861 (2012).
Brown, K. R. & Jurisica, I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 8, R95 (2007).
Bachelerie, F. et al. International Union of Pharmacology. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 66, 1–79 (2014). erratum 66, 467 (2014).
Satija, R. et al. Spatial reconstruction of the single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Wolf, F. A. et al. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Acknowledgements
We thank K. Meyer and M. Stubbington for scientific discussions; P. Porras for advice on querying the IMEx database; L. Garcia-Alonso and K. Polanski for carefully reading the manuscript; G.J. Wright, L. Wood and G. Graham for advice on protein–protein interactions; and J. Eliasova and A. Hupalowska for help with the illustrations. We are grateful to A. Lopez and YDEVS members for their help with the webserver and the implementation of the code in GitHub, as well as to all the Teichmann lab and Vento-Tormo lab members for their fruitful advice. The project was supported by Wellcome Sanger core funding (WT206194) and a Wellcome Strategic Support Science award (211276/Z/18/Z).
Author information
Authors and Affiliations
Contributions
M.E., S.A.T. and R.V.-T. conceived and developed the protocol and wrote the manuscript. M.V.-T. developed the database, implemented the code in the webserver and GitHub and contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Evangelia Petsalaki and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Vento-Tormo, R. et al. Nature 563, 347–353 (2018): https://doi.org/10.1038/s41586-018-0698-6
Stewart, B. et al. Science 365, 1461–1466 (2019): https://doi.org/10.1126/science.aat5031
Popescu, D. et al. Nature 574, 365–371 (2019): https://doi.org/10.1038/s41586-019-1652-y
Integrated supplementary information
Supplementary Figure 1 Diagram of the database structure.
a) database schema, b) protein_input/complex_input storage in the CellPhoneDB database tables. The multidata entity stores fields common to complex_input and protein_input. This makes it easier and faster for the user to perform interaction queries because interaction_table is only related to multidata_table. All non-common fields are stored in either protein_table or complex_table. Complex fields are stored in complex_composition_table. The is_complex and total_protein field are created for optimization purposes.
Supplementary Figure 2 Example of complex_input components stored in CellPhoneDB.
An example of two complex_input rows with two and four components.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Rights and permissions
About this article
Cite this article
Efremova, M., Vento-Tormo, M., Teichmann, S.A. et al. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc 15, 1484–1506 (2020). https://doi.org/10.1038/s41596-020-0292-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0292-x
This article is cited by
-
Aberrant R-loop–mediated immune evasion, cellular communication, and metabolic reprogramming affect cancer progression: a single-cell analysis
Molecular Cancer (2024)
-
Age-dependent genes in adipose stem and precursor cells affect regulation of fat cell differentiation and link aging to obesity via cellular and genetic interactions
Genome Medicine (2024)
-
Single-cell RNA sequencing of anaplastic ependymoma and H3K27M-mutant diffuse midline glioma
BMC Neurology (2024)
-
The implications of single-cell RNA-seq analysis in prostate cancer: unraveling tumor heterogeneity, therapeutic implications and pathways towards personalized therapy
Military Medical Research (2024)
-
Crosstalk among podocytes, glomerular endothelial cells and mesangial cells in diabetic kidney disease: an updated review
Cell Communication and Signaling (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.