Abstract
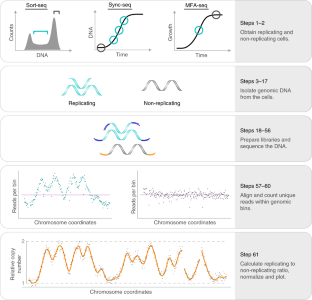
Genome replication follows a defined temporal programme that can change during cellular differentiation and disease onset. DNA replication results in an increase in DNA copy number that can be measured by high-throughput sequencing. Here we present a protocol to determine genome replication dynamics using DNA copy-number measurements. Cell populations can be obtained in three variants of the method. First, sort-seq reveals the average replication dynamics across S phase in an unperturbed cell population; FACS is used to isolate replicating and non-replicating subpopulations from asynchronous cells. Second, sync-seq measures absolute replication time at specific points during S phase using a synchronized cell population. Third, marker frequency analysis can be used to reveal the average replication dynamics using copy-number analysis in any proliferating asynchronous cell culture. These approaches have been used to reveal genome replication dynamics in prokaryotes, archaea and a wide range of eukaryotes, including yeasts and mammalian cells. We have found this approach straightforward to apply to other organisms and highlight example studies from across the three domains of life. Here we present a Saccharomyces cerevisiae version of the protocol that can be performed in 7–10 d. It requires basic molecular and cellular biology skills, as well as a basic understanding of Unix and R.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
The custom bash script required for the analysis, as well as the script to download and analyze example data, are available from GitHub (https://github.com/DNAReplicationLab/localMapper/). The R package Repliscope described here is available from CRAN (https://cran.r-project.org/web/packages/Repliscope/) and GitHub (https://github.com/DNAReplicationLab/Repliscope/). We have also provided an official Ubuntu Desktop 18.04 LTS installation disk image with all the software required for the analysis (https://ln1.path.ox.ac.uk/groups/nieduszynski/Replibuntu/Replibuntu-18.04.0-amd64.iso.gz). The code in this manuscript has been peer reviewed.
References
Fragkos, M., Ganier, O., Coulombe, P. & Mechali, M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell. Biol. 16, 360–374 (2015).
Newman, T. J., Mamun, M. A., Nieduszynski, C. A. & Blow, J. J. Replisome stall events have shaped the distribution of replication origins in the genomes of yeasts. Nucleic Acids Res. 41, 9705–9718 (2013).
Rhind, N. & Gilbert, D. M. DNA replication timing. Cold Spring Harb. Perspect. Biol. 5, a010132 (2013).
Müller, C. A. et al. The dynamics of genome replication using deep sequencing. Nucleic Acids Res. 42, e3 (2014).
Yabuki, N., Terashima, H. & Kitada, K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7, 781–789 (2002).
Raghuraman, M. K. et al. Replication dynamics of the yeast genome. Science 294, 115–121 (2001).
Peace, J. M., Villwock, S. K., Zeytounian, J. L., Gan, Y. & Aparicio, O. M. Quantitative BrdU immunoprecipitation method demonstrates that Fkh1 and Fkh2 are rate-limiting activators of replication origins that reprogram replication timing in G1 phase. Genome Res. 26, 365–375 (2016).
Sekedat, M. D. et al. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol. Syst. Biol. 6, 353 (2010).
Hawkins, M. et al. High-resolution replication profiles define the stochastic nature of genome replication initiation and termination. Cell Rep. 5, 1132–1141 (2013).
Claycomb, J. M. & Orr-Weaver, T. L. Developmental gene amplification: insights into DNA replication and gene expression. Trends Genet. 21, 149–162 (2005).
Koren, A., Soifer, I. & Barkai, N. MRC1-dependent scaling of the budding yeast DNA replication timing program. Genome Res. 20, 781–790 (2010).
Müller, C. A. & Nieduszynski, C. A. Conservation of replication timing reveals global and local regulation of replication origin activity. Genome Res. 22, 1953–1962 (2012).
Skovgaard, O., Bak, M., Lobner-Olesen, A. & Tommerup, N. Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res. 21, 1388–1393 (2011).
Hawkins, M., Malla, S., Blythe, M. J., Nieduszynski, C. A. & Allers, T. Accelerated growth in the absence of DNA replication origins. Nature 503, 544–547 (2013).
Samson, R. Y. et al. Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 3, 485–496 (2013).
Agier, N. et al. The evolution of the temporal program of genome replication. Nat. Commun. 9, 2199 (2018).
Koren, A. et al. Genetic variation in human DNA replication timing. Cell 159, 1015–1026 (2014).
Batrakou, D. G., Heron, E. D. & Nieduszynski, C. A. Rapid high-resolution measurement of DNA replication timing by droplet digital PCR. Nucleic Acids Res. 46, e112 (2018).
Shen, Y. et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science 355, eaaf4791 https://doi.org/10.1126/science.aaf4791 (2017).
Annaluru, N. et al. Total synthesis of a functional designer eukaryotic chromosome. Science 344, 55–58 (2014).
Müller, C. A. & Nieduszynski, C. A. DNA replication timing influences gene expression level. J. Cell Biol. 216, 1907–1914 (2017).
Natsume, T. et al. Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol. Cell 50, 661–674 (2013).
Kedziora, S. et al. Rif1 acts through Protein Phosphatase 1 but independent of replication timing to suppress telomere extension in budding yeast. Nucleic Acids Res. 46, 3993–4003 (2018).
Foss, E. J. et al. SIR2 suppresses replication gaps and genome instability by balancing replication between repetitive and unique sequences. Proc. Natl Acad. Sci. USA 114, 552–557 (2017).
Voichek, Y., Bar-Ziv, R. & Barkai, N. Expression homeostasis during DNA replication. Science 351, 1087–1090 (2016).
Rudolph, C. J., Upton, A. L., Stockum, A., Nieduszynski, C. A. & Lloyd, R. G. Avoiding chromosome pathology when replication forks collide. Nature 500, 608–611 (2013).
Tanaka, S., Nakato, R., Katou, Y., Shirahige, K. & Araki, H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 21, 2055–2063 (2011).
Green, B. M., Finn, K. J. & Li, J. J. Loss of DNA replication control is a potent inducer of gene amplification. Science 329, 943–946 (2010).
Tanny, R. E., MacAlpine, D. M., Blitzblau, H. G. & Bell, S. P. Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation. Mol. Biol. Cell 17, 2415–2423 (2006).
Kramara, J., Osia, B. & Malkova, A. Break-induced replication: the where, the why, and the how. Trends Genet. 34, 518–531 (2018).
Alvino, G. M. et al. Replication in hydroxyurea: it’s a matter of time. Mol. Cell. Biol. 27, 6396–6406 (2007).
Macheret, M. & Halazonetis, T. D. Monitoring early S-phase origin firing and replication fork movement by sequencing nascent DNA from synchronized cells. Nat. Protoc. 14, 51–67 (2019).
Macheret, M. & Halazonetis, T. D. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 555, 112–116 (2018).
Marchal, C. et al. Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat. Protoc. 13, 819–839 (2018).
Siow, C. C., Nieduszynska, S. R., Müller, C. A. & Nieduszynski, C. A. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res. 40, D682–D686 (2012).
Prioleau, M. N. & MacAlpine, D. M. DNA replication origins—where do we begin? Genes Dev. 30, 1683–1697 (2016).
Jinks-Robertson, S. & Klein, H. L. Ribonucleotides in DNA: hidden in plain sight. Nat. Struct. Mol. Biol. 22, 176–178 (2015).
Petryk, N. et al. Replication landscape of the human genome. Nat. Commun. 7, 10208 (2016).
McGuffee, S. R., Smith, D. J. & Whitehouse, I. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol. Cell 50, 123–135 (2013).
Feng, W. et al. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 8, 148–155 (2006).
Tiengwe, C. et al. Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Rep. 2, 185–197 (2012).
Donaldson, A. D. & Nieduszynski, C. A. Genome-wide analysis of DNA replication timing in single cells: Yes! We’re all individuals. Genome Biol. 20, 111 (2019).
Müller, C. A. et al. Capturing the dynamics of genome replication on individual ultra-long nanopore sequence reads. Nat. Methods 16, 429–436 (2019).
Haase, S. B. & Reed, S. I. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1, 132–136 (2002).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Daigaku, Y. et al. A global profile of replicative polymerase usage. Nat. Struct. Mol. Biol. 22, 192–198 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Xu, J. et al. Genome-wide identification and characterization of replication origins by deep sequencing. Genome Biol. 13, R27 (2012).
Heichinger, C., Penkett, C. J., Bahler, J. & Nurse, P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 25, 5171–5179 (2006).
Agier, N., Romano, O. M., Touzain, F., Cosentino Lagomarsino, M. & Fischer, G. The spatiotemporal program of replication in the genome of Lachancea kluyveri. Genome Biol. Evol. 5, 370–388 (2013).
Di Rienzi, S. C. et al. Maintaining replication origins in the face of genomic change. Genome Res. 22, 1940–1952 (2012).
Kim, H. S. Genome-wide function of MCM-BP in Trypanosoma brucei DNA replication and transcription. Nucleic Acids Res. 47, 634–647 (2019).
Devlin, R. et al. Mapping replication dynamics in Trypanosoma brucei reveals a link with telomere transcription and antigenic variation. Elife 5, e12765 https://doi.org/10.7554/eLife.12765 (2016).
Marques, C. A., Dickens, N. J., Paape, D., Campbell, S. J. & McCulloch, R. Genome-wide mapping reveals single-origin chromosome replication in Leishmania, a eukaryotic microbe. Genome Biol. 16, 230 (2015).
Lundgren, M., Andersson, A., Chen, L., Nilsson, P. & Bernander, R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl Acad. Sci. USA 101, 7046–7051 (2004).
Woodfine, K. et al. Replication timing of the human genome. Hum. Mol. Genet. 13, 191–202 (2004).
de Moura, A. P., Retkute, R., Hawkins, M. & Nieduszynski, C. A. Mathematical modelling of whole chromosome replication. Nucleic Acids Res. 38, 5623–5633 (2010).
Acknowledgements
We thank Amanda Williams and Becky Busby (Zoology Sequencing Facility) for help with the NextSeq 500, and Michal Maj and Line Eriksen (Sir William Dunn School Flow Cytometry Facility) for their help with FACS.
Author information
Authors and Affiliations
Contributions
All authors wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Hawkins, M. et al. Nature 503, 544–547 (2013): https://doi.org/10.1038/nature12650
Müller, C. A. et al. Nucleic Acids Res. 42, e3 (2014): https://doi.org/10.1093/nar/gkt878
Müller, C. A. & Nieduszynski, C. A. J. Cell Biol. 216, 1907–1914 (2017): https://doi.org/10.1083/jcb.201701061
Supplementary information
Supplementary Note
Shiny App walkthrough
Rights and permissions
About this article
Cite this article
Batrakou, D.G., Müller, C.A., Wilson, R.H.C. et al. DNA copy-number measurement of genome replication dynamics by high-throughput sequencing: the sort-seq, sync-seq and MFA-seq family. Nat Protoc 15, 1255–1284 (2020). https://doi.org/10.1038/s41596-019-0287-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0287-7
This article is cited by
-
The in vivo measurement of replication fork velocity and pausing by lag-time analysis
Nature Communications (2023)
-
Global early replication disrupts gene expression and chromatin conformation in a single cell cycle
Genome Biology (2022)
-
DNA read count calibration for single-molecule, long-read sequencing
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.



