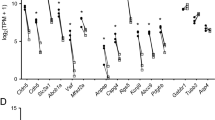
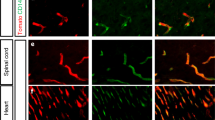
Abstract
The study of cerebral microvessels is becoming increasingly important in a wide variety of conditions, such as stroke, sepsis, traumatic brain injury and neurodegenerative diseases. However, the molecular mechanisms underlying cerebral microvascular dysfunction in these conditions are largely unknown. The molecular characterization of cerebral microvessels in experimental disease models has been hindered by the lack of a standardized method to reproducibly isolate intact cerebral microvessels with consistent cellular compositions and without the use of enzymatic digestion, which causes undesirable molecular and metabolic changes. Herein, we describe an optimized protocol for microvessel isolation from mouse brain cortex that yields microvessel fragments with consistent populations of discrete blood–brain barrier (BBB) components (endothelial cells, pericytes and astrocyte end feet) while retaining high RNA integrity and protein post-translational modifications (e.g., phosphorylation). We demonstrate that this protocol allows the quantification of changes in gene expression in a disease model (stroke) and the activation of signaling pathways in mice subjected to drug administration in vivo. We also describe the isolation of genomic DNA (gDNA) and bisulfite treatment for the assessment of DNA methylation, as well as the optimization of chromatin extraction and shearing from cortical microvessels. This optimized protocol and the described applications should improve the understanding of the molecular mechanisms governing cerebral microvascular dysfunction, which may help in the development of novel therapies for stroke and other neurologic conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. No datasets were generated or analyzed during the current study.
References
Zlokovic, B. V. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron 57, 178–201 (2008).
Shi, Y. et al. Rapid endothelial cytoskeletal reorganization enables early blood–brain barrier disruption and long-term ischaemic reperfusion brain injury. Nat. Commun. 7, 10523 (2016).
Jackman, K. et al. Progranulin deficiency promotes post-ischemic blood-brain barrier disruption. J. Neurosci. 33, 19579–19589 (2013).
Bell, R. D. et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 485, 512–516 (2012).
Montagne, A. et al. Blood-brain barrier breakdown in the aging human hippocampus. Neuron 85, 296–302 (2015).
Nation, D. A. et al. Blood-brain barrier breakdown is an early biomarker of human cognitive dysfunction. Nat. Med. 25, 270–276 (2019).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Guo, S. et al. The vasculome of the mouse brain. PLoS ONE 7, e52665 (2012).
Daneman, R. et al. The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS ONE 5, e13741 (2010).
He, L. et al. Analysis of the brain mural cell transcriptome. Sci. Rep. 6, 35108 (2016).
Paolinelli, R. et al. Wnt activation of immortalized brain endothelial cells as a tool for generating a standardized model of the blood brain barrier in vitro. PLoS ONE 8, e70233 (2013).
Ruck, T., Bittner, S., Epping, L., Herrmann, A. M. & Meuth, S. G. Isolation of primary murine brain microvascular endothelial cells. J. Vis. Exp. 2014, e52204 (2014).
Aird, W. C. Endothelial cell heterogeneity. Cold Spring Harb. Perspect. Med. 2, a006429 (2012).
Regan, E. R. & Aird, W. C. Dynamical systems approach to endothelial heterogeneity. Circ. Res. 111, 110–130 (2012).
Wu, Z., Hofman, F. M. & Zlokovic, B. V. A simple method for isolation and characterization of mouse brain microvascular endothelial cells. J. Neurosci. Methods 130, 53–63 (2003).
Munikoti, V. V., Hoang-Minh, L. B. & Ormerod, B. K. Enzymatic digestion improves the purity of harvested cerebral microvessels. J. Neurosci. Methods 207, 80–85 (2012).
Boulay, A. C., Saubamea, B., Decleves, X. & Cohen-Salmon, M. Purification of mouse brain vessels. J. Vis. Exp. 2015, e53208 (2015).
Yousif, S., Marie-Claire, C., Roux, F., Scherrmann, J. M. & Decleves, X. Expression of drug transporters at the blood-brain barrier using an optimized isolated rat brain microvessel strategy. Brain Res. 1134, 1–11 (2007).
Kim, G. S. et al. Critical role of sphingosine-1-phosphate receptor-2 in the disruption of cerebrovascular integrity in experimental stroke. Nat. Commun. 6, 7893 (2015).
Yanagida, K. et al. Size-selective opening of the blood-brain barrier by targeting endothelial sphingosine 1-phosphate receptor 1. Proc. Natl. Acad. Sci. USA 114, 4531–4536 (2017).
Lacar, B. et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 7, 11022 (2016).
Chan, Y. et al. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J. Biol. Chem. 279, 35087–35100 (2004).
Zaina, S. et al. DNA methylation map of human atherosclerosis. Circ. Cardiovasc. Genet. 7, 692–700 (2014).
Jiang, Y. Z., Manduchi, E., Stoeckert, C. J. Jr. & Davies, P. F. Arterial endothelial methylome: differential DNA methylation in athero-susceptible disturbed flow regions in vivo. BMC Genomics 16, 506 (2015).
Dunn, J. et al. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J. Clin. Invest. 124, 3187–3199 (2014).
Vanlandewijck, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018).
Hill, R. A. et al. Regional blood flow in the normal and ischemic brain is controlled by arteriolar smooth muscle cell contractility and not by capillary pericytes. Neuron 87, 95–110 (2015).
Bowyer, J. F. et al. A visual description of the dissection of the cerebral surface vasculature and associated meninges and the choroid plexus from rat brain. J. Vis. Exp. 2012, e4285 (2012).
Kono, M. et al. The sphingosine-1-phosphate receptors S1P1, S1P2, and S1P3 function coordinately during embryonic angiogenesis. J. Biol. Chem. 279, 29367–29373 (2004).
del Zoppo, G. J. & Mabuchi, T. Cerebral microvessel responses to focal ischemia. J. Cereb. Blood Flow. Metab. 23, 879–894 (2003).
Faraci, F. M. Vascular protection. Stroke 34, 327–329 (2003).
Iadecola, C. & Anrather, J. The immunology of stroke: from mechanisms to translation. Nat. Med. 17, 796–808 (2011).
Terasaki, Y. et al. Mechanisms of neurovascular dysfunction in acute ischemic brain. Curr. Med. Chem. 21, 2035–2042 (2014).
Sanchez, T. Sphingosine-1-phosphate signaling in endothelial disorders. Curr. Atheroscler. Rep. 18, 31 (2016).
Zhang, G. et al. Critical role of sphingosine-1-phosphate receptor 2 (S1PR2) in acute vascular inflammation. Blood 122, 443–455 (2013).
Fernandez-Lopez, D. et al. Blood-brain barrier permeability is increased after acute adult stroke but not neonatal stroke in the rat. J. Neurosci. 32, 9588–9600 (2012).
Bazzoni, G. & Dejana, E. Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis. Physiol. Rev. 84, 869–901 (2004).
Luissint, A. C., Artus, C., Glacial, F., Ganeshamoorthy, K. & Couraud, P. O. Tight junctions at the blood brain barrier: physiological architecture and disease-associated dysregulation. Fluids Barriers CNS 9, 23 (2012).
Mark, K. S. & Davis, T. P. Cerebral microvascular changes in permeability and tight junctions induced by hypoxia-reoxygenation. Am. J. Physiol. Heart Circ. Physiol. 282, H1485–1494 (2002).
Liu, J., Jin, X., Liu, K. J. & Liu, W. Matrix metalloproteinase-2-mediated occludin degradation and caveolin-1-mediated claudin-5 redistribution contribute to blood-brain barrier damage in early ischemic stroke stage. J. Neurosci. 32, 3044–3057 (2012).
McColl, B. W., Rothwell, N. J. & Allan, S. M. Systemic inflammation alters the kinetics of cerebrovascular tight junction disruption after experimental stroke in mice. J. Neurosci. 28, 9451–9462 (2008).
Venkataraman, K. et al. Vascular endothelium as a contributor of plasma sphingosine 1-phosphate. Circ. Res. 102, 669–676 (2008).
Sanchez, T. et al. PTEN as an effector in the signaling of antimigratory G protein-coupled receptor. Proc. Natl. Acad. Sci. USA 102, 4312–4317 (2005).
Li, Z. et al. Regulation of PTEN by Rho small GTPases. Nat. Cell Biol. 7, 399–404 (2005).
Wolfrum, S. et al. Inhibition of Rho-kinase leads to rapid activation of phosphatidylinositol 3-kinase/protein kinase Akt and cardiovascular protection. Arterioscler. Thromb. Vasc. Biol. 24, 1842–1847 (2004).
Sanchez, T. et al. Induction of vascular permeability by the sphingosine-1-phosphate receptor-2 (S1P2R) and its downstream effectors ROCK and PTEN. Arterioscler. Thromb. Vasc. Biol. 27, 1312–1318 (2007).
Ma, D. K. et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009).
Lister, R. et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013).
Nelson, J. D., Denisenko, O. & Bomsztyk, K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 (2006).
Acknowledgements
This work was supported by funds from the American Heart Association (Grant-in-Aid, 12GRNT12050110), the NIH (HL094465) and the Leducq Foundation (14CVD02) to T.S. A.I. was supported by a grant from the Roche Foundation.
Author information
Authors and Affiliations
Contributions
H.S. and T.S. designed the protocol. T.S., H.S., Y.-K.L. and H.U. modified and updated the protocol to its current state. A.I. conducted the stroke surgeries. H.U. conducted the in vivo pharmacological treatments. Y.-K.L. and H.U. optimized and conducted the molecular assays with cerebral microvessels, as well as the immunofluorescence analysis. H.U., Y.-K.L., H.S. and T.S. wrote the manuscript with contributions from all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Xavier Declèves, Sven Meuth and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
Yanagida, K. et al. Proc. Natl. Acad. Sci. USA 114, 4531–4536 (2017): https://doi.org/10.1073/pnas.1618659114
Integrated supplementary information
Supplementary Figure 1
Images of full-length blots presented in the figures.
Supplementary information
Supplementary Information
Supplementary Figure 1
Rights and permissions
About this article
Cite this article
Lee, YK., Uchida, H., Smith, H. et al. The isolation and molecular characterization of cerebral microvessels. Nat Protoc 14, 3059–3081 (2019). https://doi.org/10.1038/s41596-019-0212-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0212-0
This article is cited by
-
Impaired cerebral microvascular endothelial cells integrity due to elevated dopamine in myasthenic model
Journal of Neuroinflammation (2024)
-
Epigenetics and stroke: role of DNA methylation and effect of aging on blood–brain barrier recovery
Fluids and Barriers of the CNS (2023)
-
Comparative study of extracellular vesicles derived from mesenchymal stem cells and brain endothelial cells attenuating blood–brain barrier permeability via regulating Caveolin-1-dependent ZO-1 and Claudin-5 endocytosis in acute ischemic stroke
Journal of Nanobiotechnology (2023)
-
Spatio-temporal dynamics of microglia phenotype in human and murine cSVD: impact of acute and chronic hypertensive states
Acta Neuropathologica Communications (2023)
-
ASK1-K716R reduces neuroinflammation and white matter injury via preserving blood–brain barrier integrity after traumatic brain injury
Journal of Neuroinflammation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.