Abstract
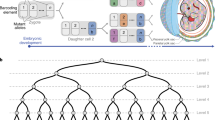
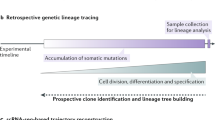
Fate mapping is a powerful genetic tool for linking stem or progenitor cells with their progeny, and hence for defining cell lineages in vivo. The resolution of fate mapping depends on the numbers of distinct markers that are introduced in the beginning into stem or progenitor cells; ideally, numbers should be sufficiently large to allow the tracing of output from individual cells. Highly diverse genetic barcodes can serve this purpose. We recently developed an endogenous genetic barcoding system, termed Polylox. In Polylox, random DNA recombination can be induced by transient activity of Cre recombinase in a 2.1-kb-long artificial recombination substrate that has been introduced into a defined locus in mice (Rosa26Polylox reporter mice). Here, we provide a step-by-step protocol for the use of Polylox, including barcode induction and estimation of induction efficiency, barcode retrieval with single-molecule real-time (SMRT) DNA sequencing followed by computational barcode identification, and the calculation of barcode-generation probabilities, which is key for estimations of single-cell labeling for a given number of stem cells. Thus, Polylox barcoding enables high-resolution fate mapping in essentially all tissues in mice for which inducible Cre driver lines are available. Alternative methods include ex vivo cell barcoding, inducible transposon insertion and CRISPR–Cas9-based barcoding; Polylox currently allows combining non-invasive and cell-type-specific labeling with high label diversity. The execution time of this protocol is ~2–3 weeks for experimental data generation and typically <2 d for computational Polylox decoding and downstream analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
All computational codes and scripts used in this protocol have been deposited in GitHub: https://github.com/hoefer-lab/RPBPBR and https://github.com/hoefer-lab/polylox. The version used for writing this protocol can be found in the Supplementary Software.
References
Snippert, H. J. & Clevers, H. Tracking adult stem cells. EMBO Rep. 12, 113–122 (2011).
Kretzschmar, K. & Watt, F. M. Lineage tracing. Cell 148, 33–45 (2012).
Woodworth, M. B., Girskis, K. M. & Walsh, C. A. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet. 18, 230–244 (2017).
Pei, W. et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460 (2017).
Sternberg, N. & Hamilton, D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 150, 467–486 (1981).
Abremski, K., Hoess, R. & Sternberg, N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell 32, 1301–1311 (1983).
Spanjaard, B. et al. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat. Biotechnol. 36, 469–473 (2018).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
McKenna, A. et al. Whole organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016).
Alemany, A., Florescu, M., Baron, C. S., Peterson-Maduro, J. & van Oudenaarden, A. Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018).
Kalhor, R. et al. Developmental barcoding of whole mouse via homing CRISPR. Science 361, eaat9804 (2018).
Spanjaard, B. & Junker, J. P. Methods for lineage tracing on the organism-wide level. Curr. Opin. Cell Biol. 49, 16–21 (2017).
Sun, J. et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014).
Rodriguez-Fraticelli, A. E. et al. Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216 (2018).
Busch, K. et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546 (2015).
Behjati, S. et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature 513, 422–425 (2014).
Lodato, M. A. et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015).
Lee-Six, H. et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018).
Weber, T. S. et al. Site-specific recombinatorics: in situ cellular barcoding with the Cre Lox system. BMC Syst. Biol. 10, 43 (2016).
Peikon, I. D., Gizatullina, D. I. & Zador, A. M. In vivo generation of DNA sequence diversity for cellular barcoding. Nucleic Acids Res. 42, e127 (2014).
Ventura, A. et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007).
Rhoads, A. & Au, K. F. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13, 278–289 (2015).
Golde, W. T., Gollobin, P. & Rodriguez, L. L. A rapid, simple, and humane method for submandibular bleeding of mice using a lancet. Lab Anim. 34, 39–43 (2005).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual. 3rd edn (Cold Spring Harbor Laboratory Press, 2001).
Buchholz, V. R. et al. Disparate individual fates compose robust CD8+ T cell immunity. Science 340, 630–635 (2013).
Perie, L. et al. Determining lineage pathways from cellular barcoding experiments. Cell Rep. 6, 617–624 (2014).
Flossdorf, M., Rossler, J., Buchholz, V. R., Busch, D. H. & Hofer, T. CD8(+) T cell diversification by asymmetric cell division. Nat. Immunol. 16, 891–893 (2015).
Cho, Y. L. et al. TCR signal quality modulates fate decisions of single CD4(+) T cells in a probabilistic manner. Cell Rep. 20, 806–818 (2017).
Xu, J. et al. Statistical inference in partially observed branching processes with application to cell lineage tracking of in vivo hematopoiesis. Preprint at https://arxiv.org/abs/1610.07550v2 (2018).
Acknowledgements
We thank C. Quedenau for invaluable help. T.H. is supported by CellNetworks, DKFZ core funding, e:Bio BMBF project FKZ 0316182B (SB-Epo), EU-FP7 ITN QuantI (317040), H2020-MSCA-ITN-2017 grant agreement 764698 and SFB 873-B11; H.-R.R. is supported by ERC Advanced Grant 742883, H2020-MSCA-ITN-2017 grant agreement 764698, SFB 873-B11 and DKFZ core funding.
Author information
Authors and Affiliations
Contributions
W.P., X.W., J.R. and T.B.F. contributed to the practical protocol. T.B.F., X.W., T.H. and H.-R.R. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information: Nature Protocols thanks Pavel Osten and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
Pei, W. et al. Nature 548, 456–460 (2017): https://doi.org/10.1038/nature23653
Supplementary information
Supplementary Software
The computational codes and scripts used in writing this protocol.
Rights and permissions
About this article
Cite this article
Pei, W., Wang, X., Rössler, J. et al. Using Cre-recombinase-driven Polylox barcoding for in vivo fate mapping in mice. Nat Protoc 14, 1820–1840 (2019). https://doi.org/10.1038/s41596-019-0163-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0163-5
This article is cited by
-
Orthogonal LoxPsym sites allow multiplexed site-specific recombination in prokaryotic and eukaryotic hosts
Nature Communications (2024)
-
Application of Lineage Tracing in Central Nervous System Development and Regeneration
Molecular Biotechnology (2023)
-
Clonal tracking in cancer and metastasis
Cancer and Metastasis Reviews (2023)
-
Mastering the use of cellular barcoding to explore cancer heterogeneity
Nature Reviews Cancer (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.