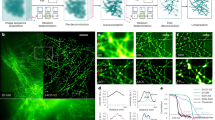
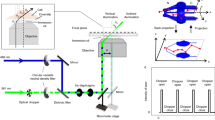
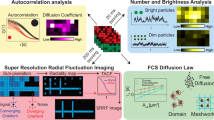
Abstract
Super-resolution microscopy techniques enable optical imaging in live cells with unprecedented spatial resolution. They unfortunately lack the temporal resolution required to directly investigate cellular dynamics at scales sufficient to measure molecular diffusion. These fast time scales are, on the other hand, routinely accessible by spectroscopic techniques such as fluorescence correlation spectroscopy (FCS). To enable the direct investigation of fast dynamics at the relevant spatial scales, FCS has been combined with super-resolution stimulated emission depletion (STED) microscopy. STED–FCS has been applied in point or scanning mode to reveal nanoscale diffusion behavior of molecules in live cells. In this protocol, we describe the technical details of performing point STED–FCS (pSTED–FCS) and scanning STED–FCS (sSTED–FCS) measurements, from calibration and sample preparation to data acquisition and analysis. We give particular emphasis to 2D diffusion dynamics in cellular membranes, using molecules tagged with organic fluorophores. These measurements can be accomplished within 4–6 h by those proficient in fluorescence imaging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The STED–FCS data are available upon request. The software used for data analysis is freely available (‘Equipment’).
References
Stefan, W. H. et al. The 2015 super-resolution microscopy roadmap. J. Phys. D 48, 443001 (2015).
Sahl, S. J., Hell, S. W. & Jakobs, S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 18, 685–701 (2017).
Elson, E. L. Fluorescence correlation spectroscopy: past, present, future. Biophys. J. 101, 2855–2870 (2011).
Sezgin, E. Super-resolution optical microscopy for studying membrane structure and dynamics. J. Phys. Condens. Matter 29, 273001 (2017).
Hell, S. W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 (1994).
Klar, T. A. & Hell, S. W. Subdiffraction resolution in far-field fluorescence microscopy. Opt. Lett. 24, 954–956 (1999).
Klar, T. A., Jakobs, S., Dyba, M., Egner, A. & Hell, S. W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA 97, 8206–8210 (2000).
Magde, D., Elson, E. L. & Webb, W. W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers 13, 29–61 (1974).
Kim, S. A., Heinze, K. G. & Schwille, P. Fluorescence correlation spectroscopy in living cells. Nat. Methods 4, 963–973 (2007).
Wawrezinieck, L., Rigneault, H., Marguet, D. & Lenne, P. F. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 89, 4029–4042 (2005).
Lenne, P. F. et al. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 25, 3245–3256 (2006).
Mueller, V. et al. STED nanoscopy reveals molecular details of cholesterol- and cytoskeleton-modulated lipid interactions in living cells. Biophys. J. 101, 1651–1660 (2011).
Schneider, F. et al. Diffusion of lipids and GPI-anchored proteins in actin-free plasma membrane vesicles measured by STED-FCS. Mol. Biol. Cell 28, 1507–1518 (2017).
Fujiwara, T. K. et al. Confined diffusion of transmembrane proteins and lipids induced by the same actin meshwork lining the plasma membrane. Mol. Biol. Cell 27, 1101–1119 (2016).
Kusumi, A., Ike, H., Nakada, C., Murase, K. & Fujiwara, T. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Semin. Immunol. 17, 3–21 (2005).
Kusumi, A. & Suzuki, K. Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim. Biophys. Acta 1746, 234–251 (2005).
Andrade, D. M. et al. Cortical actin networks induce spatio-temporal confinement of phospholipids in the plasma membrane—a minimally invasive investigation by STED-FCS. Sci. Rep. 5, 11454 (2015).
Kastrup, L., Blom, H., Eggeling, C. & Hell, S. W. Fluorescence fluctuation spectroscopy in subdiffraction focal volumes. Phys. Rev. Lett. 94, 178104 (2005).
Eggeling, C. et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162 (2009).
Clausen, M. P. et al. A straightforward approach for gated STED-FCS to investigate lipid membrane dynamics. Methods 88, 67–75 (2015).
Waithe, D. et al. Optimized processing and analysis of conventional confocal microscopy generated scanning FCS data. Methods 140-141, 62–73 (2018).
Schneider, F. et al. Nanoscale spatiotemporal diffusion modes measured by simultaneous confocal and stimulated emission depletion nanoscopy imaging. Nano Lett. 18, 4233–4240 (2018).
Vicidomini, G. et al. STED-FLCS: an advanced tool to reveal spatiotemporal heterogeneity of molecular membrane dynamics. Nano Lett. 15, 5912–5918 (2015).
Honigmann, A. et al. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat. Commun. 5, 5412–5412 (2014).
Benda, A., Ma, Y. & Gaus, K. Self-calibrated line-scan STED-FCS to quantify lipid dynamics in model and cell membranes. Biophys. J. 108, 596–609 (2015).
Wang, R. et al. A straightforward STED-background corrected fitting model for unbiased STED-FCS analyses. Methods 140-141, 212–222 (2018).
Lanzano, L. et al. Measurement of nanoscale three-dimensional diffusion in the interior of living cells by STED-FCS. Nat. Commun. 8, 65 (2017).
Koenig, M. et al. ns-time resolution for multispecies STED-FLIM and artifact free STED-FCS. in Proceedings of SPIE 9712, Multiphoton Microscopy in the Biomedical Sciences XVI (Eds. Periasamy, A., So, P.T.C. & König, K.), 97120T (2016).
Sezgin, E., Levental, I., Mayor, S. & Eggeling, C. The mystery of membrane organization: composition, regulation and physiological relevance of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374 (2017).
Bianchini, P., Cardarelli, F., Di Luca, M., Diaspro, A. & Bizzarri, R. Nanoscale protein diffusion by STED-based pair correlation analysis. PLoS ONE 9, e99619 (2014).
Hedde, P. N. et al. Stimulated emission depletion-based raster image correlation spectroscopy reveals biomolecular dynamics in live cells. Nat. Commun. 4, 2093 (2013).
Ringemann, C. et al. Exploring single-molecule dynamics with fluorescence nanoscopy. New J. Phys. 11, 103054 (2009).
Sozanski, K., Sisamakis, E., Zhang, X. & Holyst, R. Quantitative fluorescence correlation spectroscopy in three-dimensional systems under stimulated emission depletion conditions. Optica 4, 982–988 (2017).
Gao, P. & Nienhaus, G. U. Precise background subtraction in stimulated emission double depletion nanoscopy. Opt. Lett. 42, 831–834 (2017).
Chojnacki, J. et al. Envelope glycoprotein mobility on HIV-1 particles depends on the virus maturation state. Nat. Commun. 8, 545 (2017).
Urbancic, I. et al. Lipid composition but not curvature is the determinant factor for the low molecular mobility observed on the membrane of virus-like vesicles. Viruses 10, E415 (2018).
Sezgin, E. et al. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochim. Biophys. Acta 1818, 1777–1784 (2012).
Sarangi, N. K., Ayappa, K. G. & Basu, J. K. Complex dynamics at the nanoscale in simple biomembranes. Sci. Rep. 7, 11173 (2017).
Sarangi, N. K., Roobala, C. & Basu, J. K. Unraveling complex nanoscale lipid dynamics in simple model biomembranes: insights from fluorescence correlation spectroscopy in super-resolution stimulated emission depletion mode. Methods 140-141, 198–211 (2018).
Sarangi, N. K., P., I. I., Ayappa, K. G., Visweswariah, S. S. & Basu, J. K. Super-resolution stimulated emission depletion-fluorescence correlation spectroscopy reveals nanoscale membrane reorganization induced by pore-forming proteins. Langmuir 32, 9649–9657 (2016).
Honigmann, A., Mueller, V., Hell, S. W. & Eggeling, C. STED microscopy detects and quantifies liquid phase separation in lipid membranes using a new far-red emitting fluorescent phosphoglycerolipid analogue. Faraday Discuss. 161, 77–89 (2013).
Steshenko, O. et al. Reorganization of lipid diffusion by myelin basic protein as revealed by STED nanoscopy. Biophys. J. 110, 2441–2450 (2016).
Guzmán, C. et al. The efficacy of Raf kinase recruitment to the GTPase H-ras depends on H-ras membrane conformer-specific nanoclustering. J. Biol. Chem. 289, 9519–9533 (2014).
Chelladurai, R., Debnath, K., Jana, N. R. & Basu, J. K. nanoscale heterogeneities drive enhanced binding and anomalous diffusion of nanoparticles in model biomembranes. Langmuir 34, 1691–1699 (2018).
Jee, A. Y., Dutta, S., Cho, Y. K., Tlusty, T. & Granick, S. Enzyme leaps fuel antichemotaxis. Proc. Natl. Acad. Sci. USA 115, 14–18 (2018).
Zhang, X., Sisamakis, E., Sozanski, K. & Holyst, R. Nanoscopic approach to quantification of equilibrium and rate constants of complex formation at single-molecule level. J. Phys. Chem. Lett. 8, 5785–5791 (2017).
King, J. T., Yu, C., Wilson, W. L. & Granick, S. Super-resolution study of polymer mobility fluctuations near c*. ACS Nano 8, 8802–8809 (2014).
Lagerholm, B. C., Andrade, D. M., Clausen, M. P. & Eggeling, C. Convergence of lateral dynamic measurements in the plasma membrane of live cells from single particle tracking and STED-FCS. J. Phys. D Appl. Phys. 50, 063001 (2017).
Kusumi, A., Shirai, Y. M., Koyama-Honda, I., Suzuki, K. G. N. & Fujiwara, T. K. Hierarchical organization of the plasma membrane: investigations by single-molecule tracking vs. fluorescence correlation spectroscopy. FEBS Lett. 584, 1814–1823 (2010).
Reina, F. et al. Complementary studies of lipid membrane dynamics using iSCAT and super-resolved fluorescence correlation spectroscopy. J. Phys. D Appl. Phys. 51, 235401 (2018).
Sezgin, E. et al. A comparative study on fluorescent cholesterol analogs as versatile cellular reporters. J. Lipid. Res. 57, 299–309 (2016).
Humpolickova, J. et al. Probing diffusion laws within cellular membranes by Z-scan fluorescence correlation spectroscopy. Biophys. J. 91, L23–L25 (2006).
Steinberger, T., Machan, R. & Hof, M. Z-scan fluorescence correlation spectroscopy as a tool for diffusion measurements in planar lipid membranes. Methods Mol. Biol. 1076, 617–634 (2014).
Veerapathiran, S. & Wohland, T. The imaging FCS diffusion law in the presence of multiple diffusive modes. Methods 140-141, 140–150 (2018).
Jin, W., Simsek, M. F. & Pralle, A. Quantifying spatial and temporal variations of the cell membrane ultra-structure by bimFCS. Methods 140-141, 151–160 (2018).
Krieger, J. W. et al. Imaging fluorescence (cross-) correlation spectroscopy in live cells and organisms. Nat. Protoc. 10, 1948–1974 (2015).
Di Rienzo, C., Gratton, E., Beltram, F. & Cardarelli, F. Fast spatiotemporal correlation spectroscopy to determine protein lateral diffusion laws in live cell membranes. Proc. Natl. Acad. Sci. USA 110, 12307–12312 (2013).
Di Rienzo, C., Gratton, E., Beltram, F. & Cardarelli, F. Spatiotemporal fluctuation analysis: a powerful tool for the future nanoscopy of molecular processes. Biophys. J. 111, 679–685 (2016).
Digman, M. A. & Gratton, E. Imaging barriers to diffusion by pair correlation functions. Biophys. J. 97, 665–673 (2009).
Moens, P. D., Digman, M. A. & Gratton, E. Modes of diffusion of cholera toxin bound to GM1 on live cell membrane by image mean square displacement analysis. Biophys. J. 108, 1448–1458 (2015).
Malacrida, L., Hedde, P. N., Ranjit, S., Cardarelli, F. & Gratton, E. Visualization of barriers and obstacles to molecular diffusion in live cells by spatial pair-cross-correlation in two dimensions. Biomed. Opt. Express 9, 303–321 (2018).
Wenger, J. et al. Diffusion analysis within single nanometric apertures reveals the ultrafine cell membrane organization. Biophys. J. 92, 913–919 (2007).
Leutenegger, M. et al. Confining the sampling volume for Fluorescence Correlation Spectroscopy using a sub-wavelength sized aperture. Opt. Express 14, 956–969 (2006).
Regmi, R. et al. Planar optical nanoantennas resolve cholesterol-dependent nanoscale heterogeneities in the plasma membrane of living cells. Nano Lett. 17, 6295–6302 (2017).
Clausen, M. P. et al. Pathways to optical STED microscopy. NanoBioImaging 1, 1–12 (2014).
Hotta, J. et al. Spectroscopic rationale for efficient stimulated-emission depletion microscopy fluorophores. J. Am. Chem. Soc. 132, 5021–5023 (2010).
Rankin, B. R. et al. Nanoscopy in a living multicellular organism expressing GFP. Biophys. J. 100, L63–L65 (2011).
Butkevich, A. N. et al. Fluorescent rhodamines and fluorogenic carbopyronines for super-resolution STED microscopy in living cells. Angew. Chem. Int. Ed. Engl. 55, 3290–3294 (2016).
Mobarak, E. et al. How to minimize dye-induced perturbations while studying biomembrane structure and dynamics: PEG linkers as a rational alternative. Biochim. Biophys. Acta 1860, 2436–2445 (2018).
Hughes, L. D., Rawle, R. J. & Boxer, S. G. Choose your label wisely: water-soluble fluorophores often interact with lipid bilayers. PLoS ONE 9, e87649 (2014).
Moneron, G. et al. Fast STED microscopy with continuous wave fiber lasers. Opt. Express 18, 1302–1309 (2010).
Hense, A. et al. Monomeric Garnet, a far-red fluorescent protein for live-cell STED imaging. Sci. Rep. 5, 18006 (2015).
Strack, R. L. et al. A rapidly maturing far-red derivative of DsRed-Express2 for whole-cell labeling. Biochemistry 48, 8279–8281 (2009).
Morozova, K. S. et al. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys. J. 99, L13–L15 (2010).
Giepmans, B. N., Adams, S. R., Ellisman, M. H. & Tsien, R. Y. The fluorescent toolbox for assessing protein location and function. Science 312, 217–224 (2006).
Gendreizig, S., Kindermann, M. & Johnsson, K. Induced protein dimerization in vivo through covalent labeling. J. Am. Chem. Soc. 125, 14970–14971 (2003).
Stagge, F., Mitronova, G. Y., Belov, V. N., Wurm, C. A. & Jakobs, S. SNAP-, CLIP- and Halo-tag labelling of budding yeast cells. PLoS ONE 8, e78745 (2013).
Maraspini, R., Beutel, O. & Honigmann, A. Circle scanning STED fluorescence correlation spectroscopy to quantify membrane dynamics and compartmentalization. Methods 140-141, 188–197 (2018).
Waithe, D., Clausen, M. P., Sezgin, E. & Eggeling, C. FoCuS-point: software for STED fluorescence correlation and time-gated single photon counting. Bioinformatics 32, (958–960 (2015).
Muller, P., Schwille, P. & Weidemann, T. PyCorrFit-generic data evaluation for fluorescence correlation spectroscopy. Bioinformatics 30, 2532–2533 (2014).
Theer, P., Mongis, C. & Knop, M. PSFj: know your fluorescence microscope. Nat. Methods 11, 981–982 (2014).
Moffitt, J. R., Osseforth, C. & Michaelis, J. Time-gating improves the spatial resolution of STED microscopy. Opt. Express 19, 4242–4254 (2011).
Wahl, M., Gregor, I., Patting, M. & Enderlein, J. Fast calculation of fluorescence correlation data with asynchronous time-correlated single-photon counting. Opt. Express 11, 3583–3591 (2003).
Rossow, M. J., Sasaki, J. M., Digman, M. A. & Gratton, E. Raster image correlation spectroscopy in live cells. Nat. Protoc. 5, 1761–1774 (2010).
Wachsmuth, M. et al. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol. 33, 384–389 (2015).
Enderlein, J., Gregor, I., Patra, D. & Fitter, J. Art and artefacts of fluorescence correlation spectroscopy. Curr. Pharm. Biotechnol. 5, 155–161 (2004).
Bacia, K. & Schwille, P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat. Protoc. 2, 2842–2856 (2007).
Hiramoto-Yamaki, N. et al. Ultrafast diffusion of a fluorescent cholesterol analog in compartmentalized plasma membranes. Traffic 15, 583–612 (2014).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Galiani, S. et al. Strategies to maximize the performance of a STED microscope. Opt. Express 20, 7362–7374 (2012).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003).
Gautier, A. et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15, 128–136 (2008).
Los, G. V. et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008).
Acknowledgements
We thank the Wolfson Imaging Centre Oxford and the Micron Advanced Bioimaging Unit (Wellcome Trust Strategic Award 091911) for providing the microscope facility and financial support. We acknowledge funding by the Wolfson Foundation, the Medical Research Council (MRC, grant no. MC_UU_12010/unit programs G0902418 and MC_UU_12025), MRC/BBSRC/EPSRC (grant no. MR/K01577X/1), the Wellcome Trust (grant no. 104924/14/Z/14), the Deutsche Forschungsgemeinschaft (Research unit 1905 ‘Structure and function of the peroxisomal translocon’) and Oxford internal funds (John Fell Fund and EPA Cephalosporin Fund). E.S. is funded by a Newton-Katip Ҫelebi Institutional Links grant (352333122). I.U. is grateful for support from the European Commission through a Marie Skłodowska-Curie individual fellowship (707348). D.W. is funded by a URKI MRC grant (MR/S005382/1).
Author information
Authors and Affiliations
Contributions
E.S., F.S., S.G., I.U., D.W., B.C.L. and C.E. all took part in acquiring and analyzing the data and writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information: Nature Protocols thanks Rudolf Rigler and the other (anonymous) reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Eggeling, C. et al. Nature 457, 1159–1162 (2009): https://www.nature.com/articles/nature07596
Honigmann, A. et al. Nat. Commun. 5, 5412 (2014): https://www.nature.com/articles/ncomms6412
Schneider, F. et al. Mol. Biol. Cell 28, 1507–1518 (2017): https://www.molbiolcell.org/doi/10.1091/mbc.e16-07-0536
Integrated supplementary information
Supplementary Figure 1
Loading the TCSPS data.
Supplementary Figure 2
Processing the TCSPS data.
Supplementary Figure 3
Fitting the correlated data.
Supplementary Figure 4
Navigating and saving the fit results.
Supplementary Figure 5
Loading the time mode data.
Supplementary Figure 6
Processing the sSTED–FCS data.
Supplementary Figure 7
Photobleaching correction for sSTED–FCS data.
Supplementary Figure 8
Spatial and temporal cropping of sSTED–FCS data.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8
Rights and permissions
About this article
Cite this article
Sezgin, E., Schneider, F., Galiani, S. et al. Measuring nanoscale diffusion dynamics in cellular membranes with super-resolution STED–FCS. Nat Protoc 14, 1054–1083 (2019). https://doi.org/10.1038/s41596-019-0127-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0127-9
This article is cited by
-
A combination of surface-initiated controlled radical polymerization (SET-LRP) and click-chemistry for the chemical modification and fluorescent labeling of cellulose nanofibrils: STED super-resolution imaging of a single fibril and a single fibril embedded in a composite
Cellulose (2023)
-
Quantifying F-actin patches in single melanoma cells using total-internal reflection fluorescence microscopy
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.