Abstract
Morphological rearrangement of the endoplasmic reticulum (ER) is critical for metazoan mitosis. Yet, how the ER is remodeled by the mitotic signaling remains unclear. Here, we report that mitotic Aurora kinase A (AURKA) employs a small GTPase, Rab1A, to direct ER remodeling. During mitosis, AURKA phosphorylates Rab1A at Thr75. Structural analysis demonstrates that Thr75 phosphorylation renders Rab1A in a constantly active state by preventing interaction with GDP-dissociation inhibitor (GDI). Activated Rab1A is retained on the ER and induces the oligomerization of ER-shaping protein RTNs and REEPs, eventually triggering an increase of ER complexity. In various models, from Caenorhabditis elegans and Drosophila to mammals, inhibition of Rab1AThr75 phosphorylation by genetic modifications disrupts ER remodeling. Thus, our study reveals an evolutionarily conserved mechanism explaining how mitotic kinase controls ER remodeling and uncovers a critical function of Rab GTPases in metaphase.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The crystal structure of GDP-bound Rab1A-T75D is available in the Protein Data Bank under accession code PDB 7EQ2. SILAC–MS data shown in Extended Data Figure 1f are available with the manuscript. Source data are provided with this paper.
References
Champion, L., Linder, M. I. & Kutay, U. Cellular reorganization during mitotic entry. Trends Cell Biol. 27, 26–41 (2017).
Jongsma, M. L., Berlin, I. & Neefjes, J. On the move: organelle dynamics during mitosis. Trends Cell Biol. 25, 112–124 (2015).
Carlton, J. G., Jones, H. & Eggert, U. S. Membrane and organelle dynamics during cell division. Nat. Rev. Mol. Cell Biol. 21, 151–166 (2020).
Schlaitz, A. L., Thompson, J., Wong, C. C., Yates, J. R. 3rd & Heald, R. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev. Cell 26, 315–323 (2013).
Merta, H. et al. Cell cycle regulation of ER membrane biogenesis protects against chromosome missegregation. Dev. Cell 56, 3364–3379 (2021).
Smyth, J. T., Beg, A. M., Wu, S., Putney, J. W. Jr. & Rusan, N. M. Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr. Biol. 22, 1487–1493 (2012).
Nourbakhsh, K., Ferreccio, A. A., Bernard, M. J. & Yadav, S. TAOK2 is an ER-localized kinase that catalyzes the dynamic tethering of ER to microtubules. Dev. Cell 56, 3321–3333 e3325 (2021).
Lu, L., Ladinsky, M. S. & Kirchhausen, T. Cisternal organization of the endoplasmic reticulum during mitosis. Mol. Biol. Cell 20, 3471–3480 (2009).
Lu, L., Ladinsky, M. S. & Kirchhausen, T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J. Cell Biol. 194, 425–440 (2011).
Puhka, M., Joensuu, M., Vihinen, H., Belevich, I. & Jokitalo, E. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol. Biol. Cell 23, 2424–2432 (2012).
Kumar, D., Golchoubian, B., Belevich, I., Jokitalo, E. & Schlaitz, A. L. REEP3 and REEP4 determine the tubular morphology of the endoplasmic reticulum during mitosis. Mol. Biol. Cell 30, 1377–1389 (2019).
Puhka, M., Vihinen, H., Joensuu, M. & Jokitalo, E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179, 895–909 (2007).
Maiato, H., Gomes, A. M., Sousa, F. & Barisic, M. Mechanisms of chromosome congression during mitosis. Biology 6, 13 (2017).
Nigg, E. A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2, 21–32 (2001).
Wang, G., Jiang, Q. & Zhang, C. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J. Cell Sci. 127, 4111–4122 (2014).
Chin-Yo Lin et al. Peripheral Golgi protein GRASP65 is a target of mitotic polo-like kinase (Plk) and Cdc2. Proc. Natl Acad. Sci. USA 97, 12589–12594 (2000).
Sütterlin, C., Hsu, P., Mallabiabarrena, A. & Malhotra, V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell 109, 359–369 (2002).
Preisinger, C. et al. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 24, 753–765 (2005).
Kashatus, D. F. et al. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 13, 1108–1115 (2011).
Salazar-Roa, M. & Malumbres, M. Fueling the cell division cycle. Trends Cell Biol. 27, 69–81 (2017).
Hu, J. et al. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319, 1247–1250 (2008).
Voeltz, G. K., Prinz, W. A., Shibata, Y., Rist, J. M. & Rapoport, T. A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586 (2006).
Hu, J. et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561 (2009).
Orso, G. et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983 (2009).
Powers, R. E., Wang, S., Liu, T. Y. & Rapoport, T. A. Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature 543, 257–260 (2017).
Shibata, Y. et al. Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788 (2010).
Waschbusch, D. & Khan, A. R. Phosphorylation of Rab GTPases in the regulation of membrane trafficking. Traffic 21, 712–719 (2020).
Xu, L., Nagai, Y., Kajihara, Y., Ito, G. & Tomita, T. The regulation of Rab GTPases by phosphorylation. Biomolecules 11, 1340 (2021).
Eguchi, T. et al. LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc. Natl Acad. Sci. USA 115, E9115–E9124 (2018).
Hanafusa, H. et al. LRRK1 phosphorylation of Rab7 at S72 links trafficking of EGFR-containing endosomes to its effector RILP. J. Cell Sci. 132, jcs228809 (2019).
Warren, G. Membrane traffic and organelle division. Trends Biochem. Sci. 10, 439–443 (1985).
Barr, A. R. & Gergely, F. Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120, 2987–2996 (2007).
Marumoto, T., Zhang, D. & Saya, H. Aurora-A—a guardian of poles. Nat. Rev. Cancer 5, 42–50 (2005).
Yan, M. et al. Aurora-A kinase: a potent oncogene and target for cancer therapy. Med. Res. Rev. 36, 1036–1079 (2016).
Ostermeier, C. & Brunger, A. T. Structural basis of Rab effector specificity crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell 96, 363–374 (1999).
Pfeffer, S. & Aivazian, D. Targeting Rab GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 5, 886–896 (2004).
Yu, H. & Schreiber, S. L. Structure of guanine-nucleotide-exchange factor human Mss4 and identification of its Rab-interacting surface. Nature 376, 788–791 (1995).
Steger, M. et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5, e12813 (2016).
Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 (2009).
Bos, J. L., Rehmann, H. & Wittinghofer, A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 (2007).
Heo, J.-M. et al. RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK–PARKIN pathway. Sci. Adv. 4, eaav0443 (2018).
Waschbusch, D. et al. Structural basis for Rab8a recruitment of RILPL2 via LRRK2 phosphorylation of switch 2. Structure 28, 406–417 e406 (2020).
Steger, M. et al. Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. eLlie 6, e31012 (2017).
Waschbusch, D., Purlyte, E. & Khan, A. R. Dual arginine recognition of LRRK2 phosphorylated Rab GTPases. Biophys. J. 120, 1846–1855 (2021).
Gavriljuk, K., Itzen, A., Goody, R. S., Gerwert, K. & Kotting, C. Membrane extraction of Rab proteins by GDP dissociation inhibitor characterized using attenuated total reflection infrared spectroscopy. Proc. Natl Acad. Sci. USA 110, 13380–13385 (2013).
English, A. R. & Voeltz, G. K. Rab10 GTPase regulates ER dynamics and morphology. Nat. Cell Biol. 15, 169–178 (2013).
Gerondopoulos, A. et al. Rab18 and a Rab18 GEF complex are required for normal ER structure. J. Cell Biol. 205, 707–720 (2014).
Dabora, S. L. & Sheetz, M. P. The microtubule-dependent formation of a tubulovesicular network with characteristics of the ER from cultured cell extracts. Cell 54, 27–35 (1988).
Diaz, U. et al. Microtubules are necessary for proper Reticulon localization during mitosis. PLoS ONE 14, e0226327 (2019).
Dayel, M. J., Hom, E. F. Y. & Verkman, A. S. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys. J. 76, 2843–2851 (1999).
Cui-Wang, T. et al. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell 148, 309–321 (2012).
Niu, L. et al. Atlastin-mediated membrane tethering is critical for cargo mobility and exit from the endoplasmic reticulum. Proc. Natl Acad. Sci. USA 116, 14029–14038 (2019).
Hurt, C. M. et al. REEP1 and REEP2 proteins are preferentially expressed in neuronal and neuronal-like exocytotic tissues. Brain Res. 1545, 12–22 (2014).
Li, M., Liu, J. & Song, J. Nogo goes in the pure water: solution structure of Nogo-60 and design of the structured and buffer-soluble Nogo-54 for enhancing CNS regeneration. Protein Sci. 15, 1835–1841 (2006).
Shemesh, T. et al. A model for the generation and interconversion of ER morphologies. Proc. Natl Acad. Sci. USA 111, E5243–E5251 (2014).
Shibata, Y. et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283, 18892–18904 (2008).
Terasaki, M. et al. Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs. Cell 154, 285–296 (2013).
Wang, N. et al. Mechanism of membrane-curvature generation by ER-tubule shaping proteins. Nat. Commun. 12, 568 (2021).
Coppola, U., Ristoratore, F., Albalat, R. & D’Aniello, S. The evolutionary landscape of the Rab family in chordates. Cell. Mol. Life Sci. 76, 4117–4130 (2019).
Weimer, A. K., Demidov, D., Lermontova, I., Beeckman, T. & Van Damme, D. Aurora kinases throughout plant development. Trends Plant Sci. 21, 69–79 (2016).
Wu, Y., Yang, D. & Chen, G.-Y. Targeted disruption of Rab1a causes early embryonic lethality. Int. J. Mol. Med. 49, 46 (2022).
Kano, F., Tanaka, A. R., Yamauchi, S., Kondo, H. & Murata, M. Cdc2 kinase-dependent disassembly of endoplasmic reticulum (ER) exit sites inhibits ER-to-golgi vesicular transport during mitosis. Mol. Biol. Cell 15, 4289–4298 (2004).
Zaal, K. J. et al. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell 99, 589–601 (1999).
Xiang, Y., Lyu, R. & Hu, J. Oligomeric scaffolding for curvature generation by ER tubule-forming proteins. Nat. Commun. 14, 2617 (2023).
Segawa, K., Tamura, N. & Mima, J. Homotypic and heterotypic trans-assembly of human Rab-family small GTPases in reconstituted membrane tethering. J. Biol. Chem. 294, 7722–7739 (2019).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Zheng, F. et al. Nuclear AURKA acquires kinase-independent transactivating function to enhance breast cancer stem cell phenotype. Nat. Commun. 7, 10180 (2016).
Bian, X. et al. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl Acad. Sci. USA108, 3976–3981 (2011).
Yang, F. et al. Stoichiometry and regulation network of Bcl-2 family complexes quantified by live-cell FRET assay. Cell. Mol. Life Sci. 77, 2387–2406 (2020).
Wang, Q. S. et al. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 29, 68 (2018).
Zhang, W. Z. et al. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl. Sci. Tech. 30, 170 (2019).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr 66, 486–501 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr 67, 355–367 (2011).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr 66, 12–21 (2010).
Adler, J. & Parmryd, I. Colocalization analysis in fluorescence microscopy. Methods Mol. Biol. 931, 97–109 (2013).
Acknowledgements
We would like to thank H. Cheng (Peking University) and T. Tang (Chinese Academy of Sciences) for discussions, H. Li and Q. Zhang (Sun Yat-sen University) for their assistance with TEM characterization, and S. Chen, Z. Li, and S. Wu (Sun Yat-sen University Cancer Center) for technical support. We thank SYSU Instrumental Analysis and Research Center for performing MS. This work was supported by the National Key R&D Program of China (2019YFA0110300, 2017YFA0505600-04 and 2022YFA1104002 to Q.L., 2018YFA0508300 to S.G.), the National Natural Science Foundation of China (81820108024, 81630005 and 82341020 to Q.L., 82173098 to S.G., 82173367 and 81773166 to Z.W., 81972594 to M.Y., 32100589 to W.Z.), the Innovative Research Team in University of Ministry of Education of China (IRT-17R15 to Q.L.), the Natural Science Foundation of Guangdong (2016A030311038 and 2017A030313608 to Q.L., 2022A1515010915 and 2017A020215098 to Z.W., 2018A0303130299 and 2020A1515010608 to M.Y.), the Fundamental Research Funds for the Central Universities (19ykpy187 to M.Y.)., the Science and Technology Planning Project of Guangzhou (201804020044 to Q.L.), and the Cancer Innovative Research Program of Sun Yat-sen University Cancer Center (CIRP-SYSUCC-0019 to Q.L.).
Author information
Authors and Affiliations
Contributions
W.Z. and Z.Z. developed the experimental protocol; designed, performed, and analyzed the experiments; and wrote the manuscript. Y.X., D.-D.G., and J.C. designed, performed, and analyzed the experiments. Y.S. and Y.C. designed and performed the C. elegans embryo mitosis experiment. S.Z. and T.C. designed and performed the E-FRET experiment. J.X. contributed to development of the experimental protocols. B.D., D.Z., J.L., J.Z., X.L., Y.C. Y.-L.C., T.J., and C.L. contributed to the experiments. B.H., Z.L., M.Y., Z.W., B.J., and D.L. revised the experimental data and contributed to the discussion. S.G. and J.H. conceptualized, designed, and analyzed the experiments and wrote the manuscript. Q.L. directed the project, conceptualized and designed the experiments, interpreted the results, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Richard Bayliss, Blake Riggs and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Katarzyna Ciazynska and Carolina Perdigoto, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
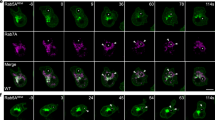
Extended Data Fig. 1 Rab1A is phosphorylated and activated during mitosis.
a, The possible phosphorylation sites of Rab1A during mitosis. b, The conserved AURKA recognition motif of Rab1A. c, Cell lysates from serum-starved (G1), double thymidine blocked (S), or nocodazole-arrested (M) MDA-MB-231 cells with or without VX680 (100 nM) treatment were analyzed by immunoblotting. d, Cell lysates were prepared at the indicated times after release from arrest with nocodazole and analyzed by immunoblotting. e, Schematic depiction of the experimental workflow of SILAC-MS analysis. Protein lysates from stable WT Flag-Rab1A or Flag-Rab1A-T75A-expressing MDA-MB-231 cells grown in ‘heavy’ and ‘light’ SILAC medium were used in anti-Flag immunoprecipitation and LC-MS/MS analyses. f, SILAC H/L ratios. The differential binding preferences (log2, y-axis) of all 63 proteins identified and quantified by LC-MS/MS (x-axis) are shown. g, Anti-Flag immunoprecipitates were prepared from MDA-MB-231 cells that stably expressed wild-type or mutant Flag-Rab1A. Immunoprecipitates and cell lysates were analyzed by immunoblotting. h, Anti-Flag immunoprecipitates were prepared from MDA-MB-231 cells that stably expressed wild-type or mutant Flag-Rab1B, Flag-Rab10, or Flag-Rab18. Immunoprecipitates and cell lysates were analyzed by immunoblotting. i, Cell lysates from MDA-MB-231 cells expressing Flag-tagged wild-type or mutant Rab1A were incubated with GTP-agarose beads in the presence or absence of GTP and the bound proteins analyzed by immunoblotting. Data shown in c, d, g, h, i are representative of two independent experiments.
Extended Data Fig. 2 Structural analysis of Rab1AThr75 phosphorylation.
a, Structural overlay of the two molecules (chains) in the asymmetric unit of the Rab1A-T75D crystal. Switch II region is specified. Note the consistent configuration of T75D in the two chains. b, Local interactions of the switch II loop (coloured magenta) in the crystal packing of Rab1A-T75D. The two crystallographic Rab1A-T75D dimers are coloured cyan and grey, respectively. NC, non-crystallographic. c, Comparison of the nucleotide binding pockets of WT Rab1A (PDB code 2FOL) and Rab1A-T75D. Note that R72 (yellow) blocks the exit of the pocket in Rab1A-T75D. d, Structural comparison between Rab1A-T75D-•GDP and Rab8A-Q67L-pT72•GTP:RILPL2 complex (PDB code 6RIR). Note the different conformations of switch II in Rab1A and Rab8A. e, Complex structures of Rab1A•GDP•AlF3 with Shigella VirA (left, 4FMB), E. Coli EspG (middle, 4FMD), and Legionella LepB (right, 4JVS). Note the different distances between Rab1A Thr75 and these GAPs.
Extended Data Fig. 3 Phosphorylation promotes Rab10 and Rab18, but not Rab1B, accumulating on the ER.
a, MDA-MB-231 cells co-expressing ER-RFP and GFP-tagged wild-type or mutant Rab1B were analyzed by confocal fluorescent microscopy. Scale bars = 20 μm. b, Colocalization of ER-RFP and GFP-tagged wild-type or mutant Rab1B was quantified in multiple cells, and evaluated by Pearson’s correlation coefficient. WT, n = 22; T72A, n = 19; T72D, n = 27. c, MDA-MB-231 cells co-expressing ER-RFP and GFP-tagged wild-type or mutant Rab10 were analyzed by confocal fluorescent microscopy. Scale bars = 20 μm. d, Colocalization of ER-RFP and GFP-tagged wild-type or mutant Rab10 was quantified in multiple cells, and evaluated by Pearson’s correlation coefficient. WT, n = 15; T73A, n = 15; T73D, n = 21. e, MDA-MB-231 cells co-expressing ER-RFP and GFP-tagged wild-type or mutant Rab18 were analyzed by confocal fluorescent microscopy. Scale bars = 20 μm. f, Colocalization of ER-RFP and GFP-tagged wild-type or mutant Rab18 was quantified in multiple cells, and evaluated by Pearson’s correlation coefficient. WT, n = 16; T72A, n = 17; T72D, n = 16. g, Whole cell lysate and ER extract from MDA-MB-231 cells were analyzed by immunoblotting. Data shown in g are representative of two independent experiments. Data from a-f were examined over three independent experiments. Data from b, d, f were analyzed using one-way Welch’s ANOVA test (P = 0.2830 in b, P < 0.0001 in d and f), and pairwise comparisons were performed using Dunnett’s T3 multiple comparisons test. Data are presented as mean ± s.e.m.
Extended Data Fig. 4 Nocodazole, but not STLC, disrupts mitotic ER morphology.
a, Schematic of living cell imaging of ER-GFP-labeled mitotic cells using three-dimensional structured illumination microscopy (3D-SIM). The length of ER elements was calculated using ImageJ software. b, Left, representative 3D-SIM images of the ER in ER-GFP-labeled MDA-MB-231 cells treated with DMSO, nocodazole, or STLC. Red boxed regions were enlarged and subjected to 3D reconstruction. Scale bars = 5 μm. Right, the length of ER elements from multiple cells in the optical SIM sections. DMSO, n = 8; Nocodazole, n = 8; STLC, n = 7. Data from b were examined over three independent experiments, and analyzed using one-way Welch’s ANOVA test (P < 0.0001), pairwise comparisons were performed using Games-Howell’s multiple comparisons test. Data are presented as mean ± s.e.m.
Extended Data Fig. 5 ER undergoes a wave of remodeling along with mitotic progression.
a, Schematic illustrating the FLIP experiments in ER-GFP-labeled mitotic cells. The repeatedly bleached region (Bleach region, circle) and fluorescence test region (Test region, square) are represented by circles and rectangles, respectively. b, c, FLIP experiments in mitotic COS-7 cells expressing ER-GFP. b, Representative images recorded by time-lapse microscopy at each time point. Scale bars = 5 μm. c, Left, average fluorescence intensity of ER-GFP from multiple cells at each time point in the test region. Right, quantification of fluorescence intensity for ER-GFP at t = 20 s derived from non-linear fitted curves. PROM, prometaphase (n = 6); META, metaphase (n = 5); ANA, anaphase (n = 5). d, Left, representative 3D-SIM images of the ER in ER-GFP-labeled mitotic COS-7 cells. Red boxed regions were enlarged and subjected to 3D reconstruction. Scale bars = 5 μm. Right, the length of ER elements from multiple cells in the optical SIM sections. PROM, n = 8; META, n = 8; ANA, n = 8. Data from b-d were examined over three independent experiments. Data from c, d were analyzed using one-way Welch’s ANOVA test (P = 0.1132 in c, P < 0.0001 in d), pairwise comparisons in c were performed using Dunnett’s T3 multiple comparisons test, pairwise comparisons in d were performed using Games-Howell’s multiple comparisons test. Data are presented as mean ± s.e.m.
Extended Data Fig. 6 AURKA-Rab1 axis enhances ER complexity during mitosis.
a, Cell lysates from shcontrol or shAURKA MDA-MB-231 cells were analyzed by immunoblotting. b, c, Doxycycline-induced shAURKA MDA-MB-231 cells were labeled with ER-GFP and used in FLIP experiments. b, Representative images recorded by time-lapse microscopy at each time point. Scale bars = 5 μm. c, Left, average fluorescence intensity for ER-GFP from multiple cells at each time point in the test region. Right, quantification of fluorescence intensity for ER-GFP at t = 20 s derived from non-linear fitted curves. shCtrl, n = 8; shAURKA, n = 7. d, Left, doxycycline-induced shAURKA MDA-MB-231 cells were labeled with ER-GFP and analyzed by 3D-SIM. Red boxed regions were enlarged and subjected to 3D reconstruction. Scale bars = 5 μm. Right, the length of ER elements from multiple cells in the optical SIM sections. shCtrl, n = 8; shAURKA, n = 8. e, Left, doxycycline-induced shAURKA MDA-MB-231 cells were visualized by thin-section electron microscopy. Scale bars = 2 μm. Right, the length of ER elements was quantified. shCtrl, n = 5; shAURKA, n = 5. f, Cell lysates from shcontrol or shRab1A MDA-MB-231 cells were analyzed by immunoblotting. g, Left, representative 3D-SIM images of the ER in ER-GFP-labeled MDA-MB-231 cells expressing control or Rab1A shRNA. Red boxed regions were enlarged and subjected to 3D reconstruction. Scale bars = 5 μm. Right, the length of ER elements from multiple cells in the optical SIM sections. shCtrl, n = 8; shRab1A#1, n = 7; shRab1A#2, n = 8. h, Left, endogenous Rab1A-knockdown MDA-MB-231 cells overexpressing Flag-tagged wild-type or mutant Rab1A were visualized by thin-section electron microscopy. Scale bars = 2 μm. Right, the length of ER elements was quantified. shRab1A+Flag-Rab1AWT, n = 5; shRab1A+Flag-Rab1A-T75A, n = 5; shRab1A+Flag-Rab1A-T75D, n = 5. Data shown in a, f are representative of three independent experiments. Data from b, c, d, e, g, h were examined over three independent experiments. Data from g, h were analyzed using one-way Welch’s ANOVA test (P < 0.0001 in g and h), and pairwise comparisons were performed using Games-Howell’s multiple comparisons test. Data from c, d, e were analyzed using two-tailed unpaired Student’s t-test. Data are presented as mean ± s.e.m.
Extended Data Fig. 7 p-Rab1AThr75 has a subtle effect on ER morphology in interphase cells.
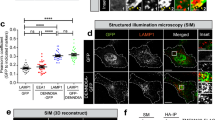
a, Left, endogenous Rab1A-knockdown MDA-MB-231 cells co-expressing mCherry-tagged wild-type or mutant Rab1A and ER-GFP were localized by N-SIM. Scale bars = 5 μm. Right, quantification of ER three-way junctions in MDA-MB-231 cells with wild-type or mutant Rab1A overexpression. WT, n = 21; T75A, n = 21; T75D, n = 21. b, Endogenous Rab1A-knockdown MDA-MB-231 cells co-expressing mCherry-tagged wild-type or mutant Rab1A and ER-GFP were treated with nocodazole and analyzed by 3D-SIM. Left, representative images of high- or low-complexity ER. Red box regions were enlarged and subjected to 3D reconstruction. Scale bars = 5 μm. Right, percentage of cells co-expressing mCherry-tagged wild-type or mutant Rab1A and ER-GFP with high-complexity ER. WT, n = 286; T75A, n = 309; T75D, n = 229. Data from a, b were examined over three independent experiments. Data from a were analyzed using one-way Welch’s ANOVA test (P = 0.5245), and pairwise comparisons were performed using Dunnett’s T3 multiple comparisons test. Data are presented as mean ± s.e.m.
Extended Data Fig. 8 Rab1A binds to ER-shaping proteins.
a-d, GST-Flag or GST-Flag-Rab1A immobilized to glutathione Sepharose 4B was used for the binding assays with the HepG2 (a), HEK293 (b, c) or Hela (d) cell lysates and the samples analyzed by immunoblotting. e, GST-Flag or GST-Flag-Rab1A immobilized to glutathione Sepharose 4B was used for the binding assays in COS-7 cell lysates and analyzed by immunoblotting. f, GST-Flag-tagged Rab1A immobilized to glutathione Sepharose 4B was used for the binding assays in COS-7 cell lysates overexpressing HA-tagged Reep1 or HA-tagged Reep5 and the samples analyzed by immunoblotting. g, Anti-GFP immunoprecipitates were prepared from MDA-MB-231 cells expressing GFP-tagged RTN4B and analyzed by immunoblotting. h, HA-tagged Reep1 bound to HA magnetic beads was used for binding assays with the COS-7 cell lysates and the samples analyzed by immunoblotting. i, j, Anti-mCherry immunoprecipitates were prepared from MDA-MB-231 cells expressing mCherry-tagged ATL1 (i) or ATL3 (j) and analyzed by immunoblotting. k, HA-tagged Reep5 bound to HA magnetic beads was used for binding assays with the COS-7 cell lysates, the samples were analyzed by immunoblotting. l, N-terminal cytosolic domains of ATL1, ATL2 and ATL3 (cyt-ATL) with HA tags were purified and incubated with GST-Flag-Rab1A in vitro for binding assays and analyzed by immunoblotting. m, HA-ATL3 was bound to anti-HA magnetic beads and then incubated with purified GST-Flag-Rab1A, the samples were analyzed by immunoblotting. n, The cytosolic loop connecting two transmembrane hairpins (TMHs) is relatively conserved in REEPs. Left, Sequence alignment of REEPs. Right, Membrane topology of REEPs. o, Binding of Rab1A to RTN peptides. Nogo-40: RIYKGVIQAIQKSDEGHPFRAYLESEVAISEELVQKYSNS (purple dots), RTN4P-1: RIYKGVIQAIQKSDEGH (pink dots), RTN4P-2: PFRAYLESEVAISEELVQKYSNS (green dots), RTN4P-3: ALGHVNCTIKELRR (deep blue dots). The protein-bound peptide fractions (bound/total) were calculated from the MST signals at each peptide concentration. p, Left, cell lysates from wild-type or ATL DKO COS-7 cells were treated with the indicated concentrations of ethylene glycol bis (EGS) and analyzed by immunoblotting with anti-Reep5 antibody. *, monomer; **, dimer; ***, trimer. Right, Reep5 dimers were quantified from three independent experiments. Data shown in a-f, h-m, p are representative of three independent experiments, and in g are from two independent experiments.
Extended Data Fig. 9 Rab1A has a subtle effect on ATL functions.
a, Rab1A has a subtle effect on ATL1, ATL2 and ATL3 GTPase activity. b, c, Left, Representative traces. Right, the gradient was fractionated into five 50 μL fractions and the reconstitution efficiency of dmATL and membrane-anchored efficiency of dmRab1 analyzed by SDS-PAGE. Data shown in b, c are representative of three independent experiments.
Extended Data Fig. 10 Phosphorylation at Thr75 promotes Rab1A interacting with ER-shaping proteins.
a, Left, anti-mCherry immunoprecipitates were prepared from MDA-MB-231 cells expressing mCherry-tagged wild-type or mutant Rab1A and analyzed by immunoblotting. Right, co-immunoprecipitated RTN4B was quantified. b, Left, anti-GFP immunoprecipitates were prepared from MDA-MB-231 cells expressing GFP-tagged wild-type or mutant Rab1A and analyzed by immunoblotting. Right, co-immunoprecipitated ATL3 was quantified. c, HA-ATL3 was bound to anti-HA magnetic beads and then incubated with purified GST-Flag-Rab1A (wild-type or mutant). The samples were analyzed by immunoblotting. d, GST-tagged wild-type or mutant Rab1A immobilized to glutathione Sepharose 4B was used for the binding assay with the COS-7 cell lysates and the samples analyzed by immunoblotting. e, mCherry-ATL3 was co-expressed with Flag-tagged WT Rab1A, Rab1A-T75A, or Rab1A-T75D in MDA-MB-231 cells. Anti-mCherry immunoprecipitation was performed and analyzed by immunoblotting. f, Left, cell lysates from COS-7 cells overexpressing RTN4B alone or co-expressing RTN4B and Rab1A (wild-type or mutant) were treated with the indicated concentrations of EGS and analyzed by immunoblotting. *, monomer; **, dimer. Right, RTN4B dimers were quantified. g, Left, cell lysates from COS-7 cells expressing wild-type or mutant Rab1A were treated with the indicated concentrations of EGS and analyzed by immunoblotting with anti-Reep5 antibody. *, monomer; **, dimer; ***, trimer. Right, Reep5 dimers were quantified. h, Left, cell lysates from COS-7 cells overexpressing Reep5 alone or co-expressing Reep5 and Rab1A (wild-type or mutant) were treated with the indicated concentrations of EGS and analyzed by immunoblotting. *, monomer; **, dimer; ***, trimer. Right, Reep5 dimers were quantified. Data shown in a-h are representative of three independent experiments. Data from a, b were analyzed using two-tailed unpaired Student’s t-test. Data are presented as mean ± s.e.m.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed western blots
Source Data Fig. 3
Unprocessed western blots
Source Data Fig. 3
Statistical source data
Source Data Fig. 4
Statistical source data
Source Data Fig. 5
Unprocessed western blots
Source Data Fig. 5
Statistical source data
Source Data Fig. 6
Unprocessed western blots
Source Data Fig. 6
Statistical source data
Source Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 1
Unprocessed western blots
Source Data Extended Data Fig. 1
Statistical source data
Source Data Extended Data Fig. 3
Unprocessed western blots
Source Data Extended Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 6
Unprocessed western blots
Source Data Extended Data Fig. 6
Statistical source data
Source Data Extended Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 8
Unprocessed western blots
Source Data Extended Data Fig. 8
Statistical source data
Source Data Extended Data Fig. 9
Unprocessed western blots
Source Data Extended Data Fig. 9
Statistical source data
Source Data Extended Data Fig. 10
Unprocessed western blots
Source Data Extended Data Fig. 10
Statistical source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, W., Zhang, Z., Xiang, Y. et al. Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis. Nat Struct Mol Biol 31, 219–231 (2024). https://doi.org/10.1038/s41594-023-01165-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-01165-7