Abstract
Protein prenylation is believed to be catalyzed by three heterodimeric enzymes: FTase, GGTase1 and GGTase2. Here we report the identification of a previously unknown human prenyltransferase complex consisting of an orphan prenyltransferase α-subunit, PTAR1, and the catalytic β-subunit of GGTase2, RabGGTB. This enzyme, which we named GGTase3, geranylgeranylates FBXL2 to allow its localization at cell membranes, where this ubiquitin ligase mediates the polyubiquitylation of membrane-anchored proteins. In cells, FBXL2 is specifically recognized by GGTase3 despite having a typical carboxy-terminal CaaX prenylation motif that is predicted to be recognized by GGTase1. Our crystal structure analysis of the full-length GGTase3–FBXL2–SKP1 complex reveals an extensive multivalent interface specifically formed between the leucine-rich repeat domain of FBXL2 and PTAR1, which unmasks the structural basis of the substrate-enzyme specificity. By uncovering a missing prenyltransferase and its unique mode of substrate recognition, our findings call for a revision of the ‘prenylation code’.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Zhang, F. L. & Casey, P. J. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269 (1996).
Wang, M. & Casey, P. J. Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 17, 110–122 (2016).
Wright, L. P. & Philips, M. R. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47, 883–891 (2006).
Ahearn, I. M., Haigis, K., Bar-Sagi, D. & Philips, M. R. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 13, 39–51 (2011).
Hougland, J. L. & Fierke, C. A. Getting a handle on protein prenylation. Nat. Chem. Biol. 5, 197–198 (2009).
Berndt, N., Hamilton, A. D. & Sebti, S. M. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer 11, 775–791 (2011).
Nguyen, U. T., Goody, R. S. & Alexandrov, K. Understanding and exploiting protein prenyltransferases. Chembiochem 11, 1194–1201 (2010).
Cox, A. D., Der, C. J., Philips, M. R. & Targeting, R. A. S. Membrane association: back to the future for anti-RAS drug discovery? Clin. Cancer Res. 21, 1819–1827 (2015).
Casey, P. J. & Seabra, M. C. Protein prenyltransferases. J. Biol. Chem. 271, 5289–5292 (1996).
Maurer-Stroh, S., Washietl, S. & Eisenhaber, F. Protein prenyltransferases. Genome Biol. 4, 212 (2003).
Lane, K. T. & Beese, L. S. Thematic review series: lipid posttranslational modifications. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J. Lipid Res. 47, 681–699 (2006).
Benetka, W., Koranda, M., Maurer-Stroh, S., Pittner, F. & Eisenhaber, F. Farnesylation or geranylgeranylation? Efficient assays for testing protein prenylation in vitro and in vivo. BMC Biochem. 7, 6 (2006).
Guo, Z. et al. Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation. EMBO J. 27, 2444–2456 (2008).
Rak, A., Pylypenko, O., Niculae, A., Goody, R. S. & Alexandrov, K. Crystallization and preliminary X-ray diffraction analysis of monoprenylated Rab7 GTPase in complex with Rab escort protein 1. J. Struct. Biol. 141, 93–95 (2003).
Pylypenko, O. et al. Structure of Rab escort protein-1 in complex with Rab geranylgeranyltransferase. Mol. Cell 11, 483–494 (2003).
James, G. L., Goldstein, J. L. & Brown, M. S. Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J. Biol. Chem. 270, 6221–6226 (1995).
Baron, R. et al. RhoB prenylation is driven by the three carboxyl-terminal amino acids of the protein: evidenced in vivo by an anti-farnesyl cysteine antibody. Proc. Natl Acad. Sci. USA 97, 11626–11631 (2000).
Carboni, J. M. et al. Farnesyltransferase inhibitors are inhibitors of Ras but not R-Ras2/TC21, transformation. Oncogene 10, 1905–1913 (1995).
Rowell, C. A., Kowalczyk, J. J., Lewis, M. D. & Garcia, A. M. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J. Biol. Chem. 272, 14093–14097 (1997).
Whyte, D. B. et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 272, 14459–14464 (1997).
Skaar, J. R., Pagan, J. K. & Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (2013).
Skaar, J. R., Pagan, J. K. & Pagano, M. SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 13, 889–903 (2014).
Yao, I. et al. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell 130, 943–957 (2007).
Koga, K., Yao, I., Setou, M. & Zhuo, M. SCRAPPER selectively contributes to spontaneous release and presynaptic long-term potentiation in the anterior cingulate cortex. J. Neurosci. 37, 3887–3895 (2017).
Chen, B. B., Coon, T. A., Glasser, J. R. & Mallampalli, R. K. Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol. Cell Biol. 31, 1905–1920 (2011).
Wang, C. et al. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18, 425–434 (2005).
Kuchay, S. et al. FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI(3)K signalling cascade. Nat. Cell Biol. 15, 472–480 (2013).
Kuchay, S. et al. PTEN counteracts FBXL2 to promote IP3R3- and Ca2+-mediated apoptosis limiting tumour growth. Nature 546, 554–558 (2017).
Kuchay, S. et al. NS5A promotes constitutive degradation of IP3R3 to counteract apoptosis induced by hepatitis C virus. Cell Rep. 25, 833–840 e833 (2018).
Lin, T. B. et al. Fbxo3-dependent Fbxl2 ubiquitination mediates neuropathic allodynia through the TRAF2/TNIK/GluR1 cascade. J. Neurosci. 35, 16545–16560 (2015).
Lai, C. Y. et al. Spinal Fbxo3-dependent Fbxl2 ubiquitination of active zone protein RIM1α mediates neuropathic allodynia through CaV2.2 activation. J. Neurosci. 36, 9722–9738 (2016).
Han, S. et al. Lipopolysaccharide primes the NALP3 inflammasome by inhibiting its ubiquitination and degradation mediated by the SCFFBXL2 E3 ligase. J. Biol. Chem. 290, 18124–18133 (2015).
Tan, M. K., Lim, H. J., Bennett, E. J., Shi, Y. & Harper, J. W. Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol. Cell 52, 9–24 (2013).
Huttlin, E. L. et al. The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 (2015).
Mellacheruvu, D. et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 (2013).
Thoma, N. H., Iakovenko, A., Goody, R. S. & Alexandrov, K. Phosphoisoprenoids modulate association of Rab geranylgeranyltransferase with REP-1. J. Biol. Chem. 276, 48637–48643 (2001).
Taylor, J. S., Reid, T. S., Terry, K. L., Casey, P. J. & Beese, L. S. Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J. 22, 5963–5974 (2003).
Furfine, E. S., Leban, J. J., Landavazo, A., Moomaw, J. F. & Casey, P. J. Protein farnesyltransferase: kinetics of farnesyl pyrophosphate binding and product release. Biochemistry 34, 6857–6862 (1995).
Park, H. W., Boduluri, S. R., Moomaw, J. F., Casey, P. J. & Beese, L. S. Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution. Science 275, 1800–1804 (1997).
Schulman, B. A. et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386 (2000).
Zheng, N. et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 (2002).
Xing, W. et al. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68 (2013).
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007).
Long, S. B., Casey, P. J. & Beese, L. S. The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 Å resolution ternary complex structures. Structure 8, 209–222 (2000).
Long, S. B., Casey, P. J. & Beese, L. S. Reaction path of protein farnesyltransferase at atomic resolution. Nature 419, 645–650 (2002).
Blomen, V. A. et al. Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015).
Otwinowski, Z. & Minor, W. in Methods in Enzymology Vol. 276 (eds C. W. Carter & R. M. Sweet) 307–326 (Academic Press, 1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002).
Acknowledgements
The authors thank G. Rona for his contribution, C. Fierke, J. Ramalho and M. Seabra for reagents, T.R. Hinds for Octet BLI analysis and M. Bergo for critically reading the manuscript. M.P. is grateful to T.M. Thor for continuous support. This work was funded by grants from the National Institutes of Health (NIH) (nos. R01-GM057587 and R01-CA076584) to M.P. and a fellowship from the T32-CA009161 (Levy) grant to A.M. M.P. and N.Z. are investigators with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
S.K. and M.P. conceived the project. S.K. designed and performed most biochemical, molecular biology and cell biology experiments. H.W. and N.Z. conceived and performed most protein purifications and all crystallization experiments. A.M., K.J. and H.H. performed some of the biochemical experiments. N.F. and M.R.P. helped with the initial microscopy experiments. M.P. and N.Z. directed and coordinated the study and oversaw the results with S.K. and H.W. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.P. is a consultant for BeyondSpring Pharmaceuticals and a member of the scientific advisory boards of CullGen, Inc. and Kymera Therapeutics. N.Z. is a member of the scientific advisory board of Kymera Therapeutics. The authors declare no other competing interests.
Additional information
Peer review information: Katarzyna Marcinkiewicz was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 PTAR1 specifically binds FBXL2 and RabGGTB.
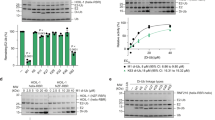
(a) IMR90 diploid fibroblasts and HEK293T cells were transfected with the indicated siRNA oligos. Seventy-two hours after siRNA transfection, cells were harvested for immunoblotting as indicated. NS: non-silencing. (b) HEK-293T cells were co-transfecetd with FLAG-tagged PTAR1 and either GFP-tagged FBXL15, GFP-tagged H-Ras, or GFP-tagged FBXL2. Twenty-four hours post-transfection, cells were harvested for immunoprecipitations and immunoblotting as indicated. (c) HEK-293T cells were transfected with the indicated siRNA oligos for 72 hours before cells were harvested for immunoblotting as indicated. NS: non-silencing, s.e.: short exposure, l.e: long exposure. (d,e) HEK-293T cells were transfected with FLAG-tagged PTAR1 and the indicated GFP-tagged substrates of FTase or GGTases. Twenty-four hours post-transfection, cells were harvested for immunoprecipitations and immunoblotting as indicated. The asterisks indicate bands visualized only after long exposures. s.e.: short exposure, l.e: long exposure. (f) HEK-293T cells were transfected with the indicated siRNA oligos for 48 hours followed by transfection with indicated cDNAs. Sixteen hours after transfection of the cDNAs, cells were lysed, and immunoprecipitations were performed with an anti-GFP antibody followed by immunoblotting as indicated. WCE: Whole cell extract. All experiments were repeated at least three times.
Supplementary Figure 2 In vitro geranylgeranylation of FBXL2 by GGTase1 and delocalization of FBXL2(C420S).
(a) Decreasing amounts of BSA (600, 300, 150, 75, or 37.5 ng) and 100 ng of either purified recombinant untagged (UT) GGTase3, or purified recombinant tagged (T; MBP-PTAR1 and GST-RabGTTB) GGTase3, were subjected to SDS-PAGE and stained with Coomassie Blue. (b) Top panel: The indicated amounts of purified FBXL2 and CDC42 were incubated with 100 ng of purified recombinant GGtase1 to carry out in vitro geranylgeranylation assay using tritiated [H3]-GGPP as described in methods. CDC42, a known GGTase1 substrate, was used as a positive control. Geranylgenranylation of FBXL2 was measured by determining the transfer of [3H]-GGPP onto purified FBXL2 and CDC42 by GGTase1 and plotted as µM/min using CPM counts. Each data point represents the GGTase1 activity (mean+/− SD)/µM/min of three technical replicates. Bottom panel: Bar graphs show in vitro geranylgeranylation assay carried out and measured as in Fig. 2b, using 10 µM of purified FBXL2, FBXW7, or CDC42 and 100 ng of purified GGTase1. Control no enzyme samples were run as biological duplicates and GGtase1 samples were run as biological triplicates. Error bar shows SEM. (c) Hela cells were transiently transfected with either GFP-tagged FBXL2 or GFP-tagged FBXL2(C420S) cDNAs. After sixteen hours, live cell imaging was carried with a LSM510 confocal microscope using a 63X objective. Images show representative frames of three independent experiments. Bar size: 10 μm.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2, Supplementary Notes 1 and 2, Supplementary Data Set 1
Rights and permissions
About this article
Cite this article
Kuchay, S., Wang, H., Marzio, A. et al. GGTase3 is a newly identified geranylgeranyltransferase targeting a ubiquitin ligase. Nat Struct Mol Biol 26, 628–636 (2019). https://doi.org/10.1038/s41594-019-0249-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0249-3
This article is cited by
-
Protein lipidation in cancer: mechanisms, dysregulation and emerging drug targets
Nature Reviews Cancer (2024)
-
Protein lipidation in health and disease: molecular basis, physiological function and pathological implication
Signal Transduction and Targeted Therapy (2024)
-
Prenylcysteine oxidase 1 like protein is required for neutrophil bactericidal activities
Nature Communications (2023)
-
Impact of a conserved N-terminal proline-rich region of the α-subunit of CAAX-prenyltransferases on their enzyme properties
Cell Communication and Signaling (2022)
-
Metabolic labeling with an alkyne probe reveals similarities and differences in the prenylomes of several brain-derived cell lines and primary cells
Scientific Reports (2021)