Abstract
Spontaneous synchronous activity is a hallmark of developing brain circuits and promotes their formation. Ex vivo, synchronous activity was shown to be orchestrated by a sparse population of highly connected GABAergic ‘hub’ neurons. The recent development of all-optical methods to record and manipulate neuronal activity in vivo now offers the unprecedented opportunity to probe the existence and function of hub cells in vivo. Using calcium imaging, connectivity analysis and holographic optical stimulation, we show that single GABAergic, but not glutamatergic, neurons influence population dynamics in the barrel cortex of non-anaesthetized mouse pups. Single GABAergic cells mainly exert an inhibitory influence on both spontaneous and sensory-evoked population bursts. Their network influence scales with their functional connectivity, with highly connected hub neurons displaying the strongest impact. We propose that hub neurons function in tailoring intrinsic cortical dynamics to external sensory inputs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data supporting the findings of this study are available from https://gitlab.com/cossartlab/. Source data are provided with this paper.
Code availability
All the codes used to extract and analyze the data illustrated in the figures are freely accessible at https://gitlab.com/cossartlab/.
References
Martini, F. J., Guillamón-Vivancos, T., Moreno-Juan, V., Valdeolmillos, M. & López-Bendito, G. Spontaneous activity in developing thalamic and cortical sensory networks. Neuron 109, 2519–2534 (2021).
Cossart, R. & Garel, S. Step by step: cells with multiple functions in cortical circuit assembly. Nat. Rev. Neurosci. 23, 395–410 (2022).
Reh, R. K. et al. Critical period regulation across multiple timescales. Proc. Natl Acad. Sci. USA 117, 23242–23251 (2020).
Luhmann, H. J. & Khazipov, R. Neuronal activity patterns in the developing barrel cortex. Neuroscience 368, 256–267 (2017).
Bonifazi, P. et al. GABAergic hub neurons orchestrate synchrony in developing hippocampal networks. Science 326, 1419–1424 (2009).
Mòdol, L. et al. Spatial embryonic origin delineates GABAergic hub neurons driving network dynamics in the developing entorhinal cortex. Cereb. Cortex 27, 4649–4661 (2017).
Feldt, S., Bonifazi, P. & Cossart, R. Dissecting functional connectivity of neuronal microcircuits: experimental and theoretical insights. Trends Neurosci. 34, 225–236 (2011).
Picardo, M. A., Guigue, P., Allene, C. & Fishell, G. Pioneer GABA cells comprise a subpopulation of hub neurons in the developing hippocampus. Neuron 71, 695–709 (2011).
Khazipov, R. et al. Early motor activity drives spindle bursts in the developing somatosensory cortex. Nature 432, 758–761 (2004).
Dooley, J. C., Glanz, R. M., Sokoloff, G. & Blumberg, M. S. Self-generated whisker movements drive state-dependent sensory input to developing barrel cortex. Curr. Biol. 30, 2404–2410 (2020).
Dzhala, V., Valeeva, G., Glykys, J., Khazipov, R. & Staley, K. Traumatic alterations in GABA signaling disrupt hippocampal network activity in the developing brain. J. Neurosci. 32, 4017–4031 (2012).
Carrillo-Reid, L., Yang, W., Miller, J.-E. K., Peterka, D. S. & Yuste, R. Imaging and optically manipulating neuronal ensembles. Annu. Rev. Biophys. 46, 271–293 (2017).
Packer, A. M., Russell, L. E., Dalgleish, H. W. P. & Häusser, M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat. Methods 12, 140–146 (2014).
Mardinly, A. R. et al. Precise multimodal optical control of neural ensemble activity. Nat. Neurosci. 21, 881–893 (2018).
Ronzitti, E., Emiliani, V. & Papagiakoumou, E. Methods for three-dimensional all-optical manipulation of neural circuits. Front. Cell. Neurosci. 12, 469 (2018).
Papagiakoumou, E., Sars, V., de, Oron, D. & Emiliani, V. Patterned two-photon illumination by spatiotemporal shaping of ultrashort pulses. Opt. Express 16, 22039 (2008).
Lutz, C. et al. Holographic photolysis of caged neurotransmitters. Nat. Methods 5, 821–827 (2008).
Landers, M. & Zeigler, H. P. Development of rodent whisking: trigeminal input and central pattern generation. Somatosens. Mot. Res. 23, 1–10 (2006).
Pnevmatikakis, E. A. et al. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299 (2016).
Rupprecht, P. et al. A database and deep learning toolbox for noise-optimized, generalized spike inference from calcium imaging. Nat. Neurosci. 24, 1324–1337 (2021).
Melzer, S. et al. Long-range-projecting GABAergic neurons modulate inhibition in hippocampus and entorhinal cortex. Science 335, 1506–1510 (2012).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2009).
Clauset, A., Shalizi, C. R. & Newman, M. E. J. Power-law distributions in empirical data. SIAM Rev. 51, 661–703 (2009).
Honey, C. J., Kötter, R., Breakspear, M. & Sporns, O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc. Natl Acad. Sci. USA 104, 10240–10245 (2007).
Sadovsky, A. J. & MacLean, J. N. Scaling of topologically similar functional modules defines mouse primary auditory and somatosensory microcircuitry. J. Neurosci. 33, 14048–14060 (2013).
Yu, S., Huang, D., Singer, W. & Nikolić, D. A small world of neuronal synchrony. Cereb. Cortex 18, 2891–2901 (2008).
Broido, A. D. & Clauset, A. Scale-free networks are rare. Nat. Commun. 10, 1017 (2019).
Das, A. & Fiete, I. R. Systematic errors in connectivity inferred from activity in strongly recurrent networks. Nat. Neurosci. 500, 1–34 (2020).
Mòdol, L. et al. Assemblies of perisomatic GABAergic neurons in the developing barrel cortex. Neuron 105, 93–105 (2019).
Golshani, P. et al. Internally mediated developmental desynchronization of neocortical network activity. J. Neurosci. 29, 10890–10899 (2009).
Sridharan, S. et al. High-performance microbial opsins for spatially and temporally precise perturbations of large neuronal networks. Neuron 110, 1139–1155 (2022).
Tuncdemir, S. N. et al. Early somatostatin interneuron connectivity mediates the maturation of deep layer cortical circuits. Neuron 89, 521–535 (2016).
Marques-Smith, A. et al. A transient translaminar GABAergic interneuron circuit connects thalamocortical recipient layers in neonatal somatosensory cortex. Neuron 89, 536–549 (2016).
Fogarty, M. et al. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 27, 10935–10946 (2007).
Chettih, S. N. & Harvey, C. D. Single-neuron perturbations reveal feature-specific competition in V1. Nature 567, 334–340 (2019).
Carrillo-Reid, L. & Yuste, R. Playing the piano with the cortex: role of neuronal ensembles and pattern completion in perception and behavior. Curr. Opin. Neurobiol. 64, 89–95 (2020).
Carrillo-Reid, L., Han, S., Yang, W., Akrouh, A. & Yuste, R. Controlling visually guided behavior by holographic recalling of cortical ensembles. Cell 178, 447–457 (2019).
Marshel, J. H. et al. Cortical layer-specific critical dynamics triggering perception. Science 365, eaaw5202 (2019).
Rickgauer, J. P., Deisseroth, K. & Tank, D. W. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat. Neurosci. 17, 1816–1824 (2014).
Robinson, N. T. M. et al. Targeted activation of hippocampal place cells drives memory-guided spatial behavior. Cell 183, 2041–2042 (2020).
Okada, T. et al. Pain induces stable, active microcircuits in the somatosensory cortex that provide a therapeutic target. Sci. Adv. 7, eabd8261 (2021).
Wrosch, J. K. et al. Rewiring of neuronal networks during synaptic silencing. Sci. Rep. 7, 11724 (2017).
Bocchio, M. et al. Hippocampal hub neurons maintain distinct connectivity throughout their lifetime. Nat. Commun. 11, 4559 (2020).
Kaiser, M. Mechanisms of connectome development. Trends Cogn. Sci. 21, 703–717 (2017).
Hu, J. S., Vogt, D., Sandberg, M. & Rubenstein, J. L. Cortical interneuron development: a tale of time and space. Development 144, 3867–3878 (2017).
Wang, C.-Z. et al. Early-generated interneurons regulate neuronal circuit formation during early postnatal development. eLife 8, 333 (2019).
García, N. V. D. M., Karayannis, T. & Fishell, G. Neuronal activity is required for the development of specific cortical interneuron subtypes. Nature 472, 351–355 (2011).
Luccioli, S. et al. Modeling driver cells in developing neuronal networks. PLoS Comput. Biol. 14, e1006551 (2018).
Kirmse, K. et al. GABA depolarizes immature neurons and inhibits network activity in the neonatal neocortex in vivo. Nat. Commun. 6, 7750 (2015).
Murata, Y. & Colonnese, M. T. GABAergic interneurons excite neonatal hippocampus in vivo. Sci. Adv. 6, eaba1430 (2020).
Steinmetz, N. A. et al. Aberrant cortical activity in multiple GCaMP6-expressing transgenic mouse lines. eNeuro 4, ENEURO.0207–17.2017 (2017).
Golshani, P. & Portera-Cailliau, C. In vivo 2-photon calcium imaging in layer 2/3 of mice. J. Vis. Exp. 681 (2008).
Chaigneau, E. et al. Two-photon holographic stimulation of ReaChR. Front. Cell. Neurosci. 10, 234 (2016).
Ronzitti, E. et al. Submillisecond optogenetic control of neuronal firing with two-photon holographic photoactivation of Chronos. J. Neurosci. 37, 10679–10689 (2017).
Chen, I.-W. et al. In vivo sub-millisecond two-photon optogenetics with temporally focused patterned light. J. Neurosci. 39, 3484–3497 (2019).
Hernandez, O., Guillon, M., Papagiakoumou, E. & Emiliani, V. Zero-order suppression for two-photon holographic excitation. Opt. Lett. 39, 5953–5956 (2014).
Pnevmatikakis, E. A. & Giovannucci, A. NoRMCorre: an online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94 (2017).
Guizar-Sicairos, M., Thurman, S. T. & Fienup, J. R. Efficient subpixel image registration algorithms. Opt. Lett. 33, 156–158 (2008).
Rubinov, M. & Sporns, O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069 (2010).
Gillespie, C. S. Fitting heavy tailed distributions: the poweRlaw package. J. Stat. Softw. 64, 1–16 (2015).
Rodrigues, F. A., Peron, T. K. D., Ji, P. & Kurths, J. The Kuramoto model in complex networks. Phys. Rep. 610, 1–98 (2016).
Acknowledgements
We thank C. Dussaux and A. Barrat for helpful discussions on previous versions of the manuscript and constructive feedback. We thank L. Cagnacci, C. Pauchet-Lopez, F. Michel (InMAGIC imaging facility) and INMED’s animal facility technicians for excellent technical support. We are grateful to V. Crépel and A. Represa for sharing equipment. We thank E. Papagiakoumou for the helpful discussion on the design of the microscope setup. We also thank all Cossart laboratory members for useful comments on this manuscript. This work was supported by the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement nos. 646925 and 951330), by the Fondation Bettencourt Schueller, by the Fondation Roger de Spoelberch and by the French National Research Agency (grant agreement no. ANR-14-CE13-0016). A.B. and R.C. are supported by CNRS. E.L. is supported by the Ministère de l’Enseignement Superieur et de la Recherche. E.R. received funding from the European Research Council Synergy Grant scheme (Helmholtz, ERC grant agreement no. 610110). V.E. acknowledges the Fondation Bettencourt Schueller (Prix Coups d’Elan pour la Recherche Française) and the ERC Advanced Grant HOLOVIS (ERC-2019-AdG, award no. 885090).
Author information
Authors and Affiliations
Contributions
L.M., Y.B., T.T. and R.C. designed the research. L.M., Y.B., T.T., E.L., S.B., A.B., A.V., R.D., M.A.P. and J.-C.P. performed experiments. L.M., Y.B. and D.A. analyzed data. L.M., Y.B., D.A. and R.C. wrote the paper. E.R. designed the optical system for holographic stimulations, with V.E., T.T., R.S., I.B. and S.B. V.D. conceived and developed the Wavefront Designer IV software, with V.E. H.A. provided the opsin viruses. R.C. conceived and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Peer review
Peer review information
Nature Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1
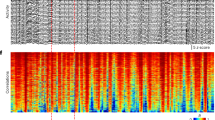
a, Representative rasterplot of the onsets of calcium events onsets occurring in imaged neurons within an imaging session at p8; all cells are labeled using viral expression of GCaMP6s (AAV1-Syn-GCaMP6s); (b) the corresponding sum of active contours as a function of time is plotted in the histogram below; red dotted line indicates the number of coactive neurons above which synchronous activity exceeds chance level (99th percentile) and black triangles indicate detected Spontaneous Calcium Events. c, Box plot indicating number of imaged cells at different developmental stages. One-way ANOVA: F(2,40) = 2.29; p = n.s. n = 12 FOVs p5-6 (3 females and 2 males); n = 23 FOVs p7-9 (4 females and 5 males); n = 8 FOVs p10-11 (4 females and 3 males). Each dot represents a FOV. d, Upper panels, z projection of in vivo GCaMP6s and tdTomato (Ai14 + GABAergic cells) expression. Scale bar 100 μm. Lower panels show viral expression at p8 of AAV1GCaMP6s in a coronal section (70 μm) of a pup injected at p0. e Left panel: Box and whiskers indicates the fraction of GABA neurons (expressing tdTomato) among the active cell population as a function of age. No differences were observed between ages: p5-6 = 18 ± 2.7 (n = 8 FOVs); p7-9 = 17 ± 3.2 (n = 14 FOVs); p10-11 = 17 ± 3.5 (n = 4 FOVs). Each dot represent a FOV. Right panel: Bar plot show the percentage of interneurons expressing GCaMP6s (n = 8 pups, 5 females and 3 males). Each dot represents a pup. f, Distribution of the median % output (1) and input (2) connectivity degree as a function of inferred spikes. Correlation with the % of Output connectivity degree: y = 0.02797*x + 3.042; Correlation with the % of Input connectivity degree: y = 0.04952*x + 2.117. Data in panel c and e (left panel) is presented as Median and Interquartile range. Data in panel e (right panel) is given as mean ± SEM.
Extended Data Fig. 2
a, Comparison of three different methods for calculating functional connectivity. In the diagonal: Distribution of out-degree obtained via Functional Connectivity calculated using 1) binary representation of the connectivity matrix as in main text. 2) weighted connectivity matrix, i.e, same as 1) but the value (i,j) of the connectivity matrix is the strength of the connection extracted as the maximum value of normalized crosscorrelation within the +/−4 frames between spike train i and j. 3) Connectivity matrix with the entry (i,j) as the Maximum value of transfer entropy between spike train i and j in the same +/−4 frames. While we only expect to obtain heavy tail distributions when quantifying the degree of the binary connectivity matrix -only measure that uses whole numbers-, the degrees calculated with all methods follow a similar degree rank, as seen by the positively correlated scatter plots of the degree values using the three methods (off- diagonal plots). b, Heatmap representing the average correlation between the degrees calculated using the three methods described above across the n = 23 FOVs which corresponds to p7 up to p9.
Extended Data Fig. 3
a, Box plots indicate number of recorder GCaMP6s positive cells as a function of age. One-way ANOVA: F(2,25) = 0.68; p = .n.s. n = 9 FOVs at p4-6 (3 females and 1 male); n = 12 FOVs at p7-9 (1 female and 2 males); n = 8 FOVs at p10-11 (2 females and 1). Each dot represents a FOV. Data are given as median and interquartile range. b, Distribution of the functional output (1) and input (2) connectivity (fraction of active) of all imaged GABA neurons (n = 4740) in the three age groups. The data is best fitted using a log normal distribution (see Methods). Arrows and numbers in pink indicate the connectivity of HC cells (95th percentile) for OUTPUT (28.7%) and INPUT (30.4%). c, Violin plots showing differences in the anatomical distance between HCout_global(1) and HCin_global (2) GABAergic cells (pink), as well as HC cells and non-HC cells (purple) and bet non-HC cells (blue). (1) Output connectivity: One-way ANOVA: F(2, 2997) = 298.0; p < 0.0001. Tukey’s (two-sided) post hoc comparison indicates differences between the distance of HC to HC vs. Non- HC to no HC (p = 1.45e-90) and HC to HC vs HC to non-HC (p = 1.05e-97). (2) Input connectivity: One-way ANOVA: F(2, 2997) = 35.47; p < 0.0001. Tukey’s (two-sided) post hoc comparison indicates differences between the anatomical distance between HC cells vs HC and non-HC cells (p = 1361e-09), HC cells vs HC to non-HC cells (p = 396e-18), and between the distance of Non-HC (among themselves) and the distance of HC to Non-HC cells (p = 0.01). Data is panels a and c is presented as Median and interquartile range. Box and whiskers statistics correspond to the analysis performed with the mean value of each animal across all recorded FOVs. Each data point represent a FOV.
Extended Data Fig. 4
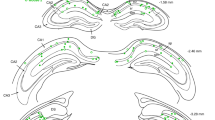
a, Representative photomicrograph showing AAV1.hSyn.GCaMP6s and AAV9.CAG.DIO.ChroME-ST.mRuby3 injections spread at P8, in GAD67Cre/+ mice (sections at 70 μm). Scale bar: 1 mm. b, Representative example and detail of AAVs infection in a GAD67Cre/+ pup injected at birth (sections at 70 μm). Scale bar: 100 μm and 50 μm. Quantification of the fraction of cells expressing ChroME and GCaMP6s. Only 41 ± 5% of the cells expressing ChroME (mRuby3) also expressed GCaMP6s (n = 5 pups (3 females and 2 males), 17 FOVs). c, Representative example and detail of AAVs infection in a representative Emx1 Cre pup injected at birth (sections at 70 μm). Scale bar :100 μm and 50 μm. Quantification of the fraction of cells expressing ChroME and GCaMP6s. 80 ± 4% of the cells expressing ChroME (mRuby3) expressed GCaMP6s (n = 3 pups (2 females and 1 male), 8 FOVs). Each dot represents an imaging session. Data are given as mean ± SEM.
Extended data Fig. 5
a, Schematic representation of the photostimulation protocol (blue light, 470 nm) used in slices of L2/3 barrel cortex (p7-9). Scale bar 20 μm b, Patched ChroME-expressing cell from a GAD67Cre mouse pup injected with AAV9.CAG.DIO.ChroME-ST.mRuby3 at birth (n = 10 cells, 4 pups, 3 females and 1 male). c, Representative spikes evoked, in ChroME-expressing GABA cells, by trains of 0.1, 0.2, and 0.5 mW/mm2 light pulses delivered at 10 (blue) and 40 Hz (pink). d, Number of action potentials (AP) in ChroME-expressing GABA cells during 10 (blue) and 40 Hz (pink) light stimulation trains as a function of LED power. Two-sided Friedman test: p < 0.0001. Dunn’s post hoc comparison indicates differences at 10 Hz between 0.1 vs. 0.2 (p = 0.015) and 0.1 vs. 0.5 (p = 0.0003) and differences at 40 Hz between 0.1 vs. 0.2 (p = 0.0304) and 0.1 vs. 0.5 (p = 0.0001) at 40 Hz. e, Delay measured in ChroME-expressing GABA cells for 10 (blue) and 40 Hz (pink) light stimulation trains at 0.2 and 0.5 mW/mm2. Wilcoxon (two-sided) test (p = 0.015, p = 0.007). f, Spike amplitude measured in ChroME-expressing GABA cells for 10 and 40 Hz stimulation trains at 0.2 and 0.5 mW/mm2. g, Patched ChroME-expressing cell from a Emx1 Cre/+ mouse pup injected with AAV9.CAG.DIO.ChroME-ST.mRuby3 at birth (n = 8 cells, 5 pups, 3 females and 2 males). Scale bar 20 μm. h, Representative spikes evoked, in ChroME-expressing glutamatergic cells, by trains of 0.1, 0.2, and 0.5 mW/mm2 light pulses delivered 10 (blue traces) and 40 Hz (pink traces). i, Number of AP in ChroME-expressing glutamatergic cells during 10 (blue) and 40 Hz (pink) light stimulation trains as a function of LED density power. Two-sided Friedman test: p < 0.0001. Dunn’s post hoc comparison indicates differences at 10 and 40 Hz between 0.1 vs. 0.5 (p = 0.0009) j, Delay measured in ChroME-expressing glutamatergic cells for 10 (blue) and 40 Hz (pink) light stimulation trains at 0.2 and 0.5 mW/mm2. Wilcoxon test (10 Hz, p = 0.015, 40 Hz, p = 0.007). k, Spike amplitude measured in ChroME- expressing glutamatergic cells for 10 and 40 Hz stimulation trains at 0.2 and 0.5 mW/mm2. Each dot represents a cell. Data in panels d and I is given as mean and −/+SEM. Data in panels e, f, j, and k are given as median and interquartile range.
Extended Data Fig. 6
a, GABA cells response to holographic stimulation. DF/F cell responses to light pulses. Panels indicate the cell response to holographic stimulation (n = 26 GABAergic cells). Grey lines correspond to a single response and black lines correspond to the averaged cell response. Each panel correspond to a single GABA cell stimulated. Responses were normalized by the pooled mean and standard deviation of the baseline (10 frames before stimulation started). b, Glutamatergic cells response to holographic stimulation. DF/F cell responses to light pulses. Panels indicate the cell response to holographic stimulation (n = 28 Glutamatergic cells). Grey lines correspond to a single response and black lines correspond to the averaged cell response. Each panel correspond to a single glutamatergic cell stimulated. Responses were normalized by the pooled mean and standard deviation of the baseline (10 frames before stimulation started).
Extended Data Fig. 7 Light pulses do not induce changes in network activity.
a–d. Example of a HC GCaMP6s positive and ChroME-ST negative stimulated cell. a, Probability distribution of cells functional output links (%). Red arrow indicates the output link connectivity of the stimulated cell. b, Correlation image of an imaging session and functional output links of the stimulated cell. c, Averaged fluorescent calcium event triggered in response to photostimulation (10 pulses, 10hz, 10 ms). Shadow indicates SEM. Data is given as Mean and −/+ SEM d, Raster plot of active cells and stimulation pulses. Grey stars indicate detected network events. Scale 5 cells. e & f, Changes in the median order parameter between Baseline, Stimulation (Stim) and Post-Stimulation (Post) for all stimulated GABA (e) (n = 11 pups, 7 females and 4 males, 26 stimulated cells) (two-sided t-test; Pre-Stim: p = 1.13e-4; Pre-Post: p = 8.21e-4; Stim-Post: p = 0.004) and glutamatergic (f) cells (n = 8 pups, 28 stimulated cells) (two-sided t-test; Pre-Stim: p = 0.01; Pre-Post: p = 0.002; Stim-Post: p = 0.71). Each dot represents a FOV. Data are given by Median and interquartile range.
Extended Data Fig. 8
a and e, Left: Schematic representation of P8–P10 GtACR1-positive cells recorded during photostimulation (blue light, 470 nm) in the L2/3 of S1BF (n = 10 cells, 3 pups, 2 females and 1 male). Right: Photocurrent measured during the stimulation protocol for different LED power (0, 0.1, 0.2, 0.3, 0.4 and 0.5 mW/mm2). b, Representative spikes evoked in GtACR1- expressing Lhx6 cells, with a single light pulse (1 s) delivered at 0, 0.1, 0.2, and 0.5 mW/mm2. c, Number of AP in GtACR1-expressing Lhx6 cells during a single pulse (1 s) as a function of LED power. Two-sided Friedman test: p = 0.07. d, Delay measured from the onset of the light pulse to the following spike in GtACR1-expressing Lhx6 cells during the application of a 1 s light pulse at 0, 0.1, 0.2 and 0.5 mW/mm2. Two-sided Friedman test: p = 0.001. Dunn’s post hoc comparison indicates differences between 0 vs. 0.2 (p < 0.05). f, Representative spikes evoked, in GtACR1-expressing Lhx6 cells, by 1 s trains of 0, 0.1, 0.2, and 0.5 mW/mm2 light pulses (10 ms) delivered at 10 Hz. g, Number of AP in GtACR1-expressing Lhx6 cells during 10 Hz light stimulation trains as a function of LED power. Two-sided Friedman test: p < 0.0001. Dunn’s post hoc comparison indicates differences at 10 Hz between 0 vs. 0.2 (p = 0.0006); 0 vs. 0.5 (p < 0.0001) and 0.1 vs. 0.5 (p = 0.014). h, Delay measured from the onset of the light pulse to the following spike in GtACR1-expressing Lhx6 cells during 10 Hz light stimulation trains at 0, 0.1, 0.2 and 0.5 mW/mm2. Friedman test: p < 0.0001. Dunn’s post hoc comparison indicates differences at 10 Hz between 0 vs. 0.2 (p = 0.0004) and 0 vs. 0.5 (p = 0.0002). In c, d, g and h, each light grey line represents a photoinhibited cell and dark lines represents median and interquartile range.
Extended data Fig. 9 DF/F cell responses to light pulses.
Panels indicate the cell response to holographic stimulation (n = 37). Grey lines correspond to a single response and black lines correspond to the averaged cell response. Each panel correspond to a single cell stimulated. Responses were normalized by the pooled mean and standard deviation of the baseline (10 frames before stimulation started). Cells that did not had a time locked response to WS were excluded from the analysis (n = 6 cells).
Extended Data Fig. 10
a, Distribution of the correlation coefficients per FOV of the cells firing activity during WS relative to control (no WS) and the In and Out degree of imaged cells. We used four different measures to characterize activity during calcium imaging (average df/f, spike frequency, average raw calcium traces and frequency of calcium onsets global activity). For all measures we found a significant relationship between differences in calcium activity during control and WS and the connectivity of the imaged cells. The amount of FOVs for which this relationship is significant is higher for In than Out degree. b, Correlation plot indicating that the cells with a high correlation coefficient between In degree and the cell firing within WS (Two-sided t-test: p < 0.001), display lower correlation with their Out degree.
Supplementary information
Supplementary Video 1
Supplementary Video 1: Example movie showing the impact on neuronal dynamics of holographic activation of a single HC GABAergic cell expressing ST-Chrome, as illustrated in Fig. 3a. Movie shows the fluorescent signal from cells expressing GCaMP6s (p8 pup imaged at 2.7 Hz, with an FOV of 350 × 350 μm2). Three time periods were recorded consecutively corresponding to baseline (1,000 frames (5 min)), stimulation (300 frames (1.5 min)) and post-stimulation (360 frames (1.8 min)) periods as shown in Fig. 4d,e. Light artifacts resulting from holographic stimulation can be observed during the stimulation period.
Supplementary Video 2
Supplementary Video 2: Example movie showing the impact on neuronal dynamics of holographic activation of a single HC glutamatergic cell expressing ST-Chrome, as illustrated in Fig. 3b. Movie shows the fluorescent signal from cells expressing GCaMP6s (p8 pup imaged at 2.7 Hz, with an FOV of 350 × 350 μm2). Three time periods were recorded consecutively corresponding to baseline (1,000 frames (5 min)), stimulation (300 frames (1.5 min)) and post-stimulation (360 frames (1.8 min)) periods as shown in Fig. 3d,e. Light artifacts resulting from holographic stimulation are visible during the stimulation period.
Source data
Source Data Fig. 1
Source Data for Fig. 1. Contains: Images: Fig. 1e, Connectivity Graphs. Data: Fig. 1c,Connection Density. Figure 1d, LCC. Figure 1f, Connectivity Distribution.
Source Data Fig. 2
Source Data for Fig. 2. Contains: Images: Fig. 2a, Connectivity Contour Map. Figure 2d, Connectivity Graphs. Data:Fig. 2b, Connectivity Comparison. Figure 2e, Connection Density. Figure 2f, LCC. Figure 2g, Comparison Highly Connected Cells. Figure 2j, Connection Density. Figure 2k, LCC. Figure 2l, Comparison Highly Connected Cells.
Source Data Fig. 3
Source Data for Fig. 3. Contains: Images: Fig. 3a2, Correlation Image Fig. 3a3, Binarized Spike Raster. Fig. 3a5, Cell Cross-Correlation. Fig. 3b2, Correlation Image. Fig. 3b3, Binarized Spike Raster. Fig. 3b5, Cell Cross-Correlation. Data: Fig. 3c, Comparison Cell Cross-Correlation.
Source Data Fig. 4
Source Data for Fig. 4. Contains: Images:Fig. 4b, Binarized Spike Raster. Figure 4e, Cell Responses. Data:Fig. 4c, Comparison Correlation in/out degree. Figure 4f, Max PSTH and max Order Parameter.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bollmann, Y., Modol, L., Tressard, T. et al. Prominent in vivo influence of single interneurons in the developing barrel cortex. Nat Neurosci 26, 1555–1565 (2023). https://doi.org/10.1038/s41593-023-01405-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-023-01405-5