Abstract
The incorporation of new information into the hippocampal network is likely to be constrained by its innate architecture and internally generated activity patterns. However, the origin, organization and consequences of such patterns remain poorly understood. In the present study we show that hippocampal network dynamics are affected by sequential neurogenesis. We birthdated CA1 pyramidal neurons with in utero electroporation over 4 embryonic days, encompassing the peak of hippocampal neurogenesis, and compared their functional features in freely moving adult mice. Neurons of the same birthdate displayed distinct connectivity, coactivity across brain states and assembly dynamics. Same-birthdate neurons exhibited overlapping spatial representations, which were maintained across different environments. Overall, the wiring and functional features of CA1 pyramidal neurons reflected a combination of birthdate and the rate of neurogenesis. These observations demonstrate that sequential neurogenesis during embryonic development shapes the preconfigured forms of adult network dynamics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The dataset generated for the present study has been made publicly available in the Buzsáki lab repository (https://buzsakilab.nyumc.org/datasets).
Code availability
Customized MATLAB scripts are available at https://github.com/buzsakilab/buzcode and https://github.com/rhuszar.
References
Amaral, D., & Lavenex, P. in The Hippocampus Book ( eds P. Andersen et al.) Hippocampal Neuroanatomy (Oxford University Press, 2007).
O’keefe, J. & Nadel, L. The Hippocampus as a Cognitive Map (Oxford Univ. Press, 1978).
Redish, A. D. et al. Independence of firing correlates of anatomically proximate hippocampal pyramidal cells. J. Neurosci. 21, RC134–RC134 (2001).
Bittner, K. C. et al. Conjunctive input processing drives feature selectivity in hippocampal CA1 neurons. Nat. Neurosci. 18, 1133–1142 (2015).
Muller, R. U., Stead, M. & Pach, J. The hippocampus as a cognitive graph. J. Gen. Physiol. 107, 663–694 (1996).
Treves, A. & Rolls, E. T. Computational analysis of the role of the hippocampus in memory. Hippocampus 4, 374–391 (1994).
Slomianka, L., Amrein, I., Knuesel, I., Sørensen, J. C. & Wolfer, D. P. Hippocampal pyramidal cells: the reemergence of cortical lamination. Brain Struct. Funct. 216, 301–317 (2011).
Mizuseki, K., Diba, K., Pastalkova, E. & Buzsáki, G. Hippocampal CA1 pyramidal cells form functionally distinct sublayers. Nat. Neurosci. 14, 1174–1181 (2011).
Lee, S.-H. et al. Parvalbumin-positive basket cells differentiate among hippocampal pyramidal cells. Neuron 82, 1129–1144 (2014).
Valero, M. et al. Determinants of different deep and superficial CA1 pyramidal cell dynamics during sharp-wave ripples. Nat. Neurosci. 18, 1281–1290 (2015).
Cembrowski, M. S. et al. Spatial gene-expression gradients underlie prominent heterogeneity of CA1 pyramidal neurons. Neuron 89, 351–368 (2016).
Masurkar, A. V. et al. Medial and lateral entorhinal cortex differentially excite deep versus superficial CA1 pyramidal neurons. Cell Rep. 18, 148–160 (2017).
Navas-Olive, A. et al. Multimodal determinants of phase-locked dynamics across deep-superficial hippocampal sublayers during theta oscillations. Nat. Commun. 11, 2217 (2020).
Fattahi, M., Sharif, F., Geiller, T. & Royer, S. Differential representation of landmark and self-motion information along the CA1 radial axis: self-motion generated place fields shift toward landmarks during septal inactivation. J. Neurosci. 38, 6766–6778 (2018).
Sharif, F., Tayebi, B., Buzsáki, G., Royer, S. & Fernandez-Ruiz, A. Subcircuits of deep and superficial CA1 place cells support efficient spatial coding across heterogeneous environments. Neuron 109, 363–376 (2021).
Lee, J. S., Briguglio, J. J., Cohen, J. D., Romani, S. & Lee, A. K. The statistical structure of the hippocampal code for space as a function of time, context, and value. Cell 183, 620–635 (2020).
Lee, D., Lin, B.-J. & Lee, A. K. Hippocampal place fields emerge upon single-cell manipulation of excitability during behavior. Science 337, 849–853 (2012).
Dragoi, G. & Tonegawa, S. Preplay of future place cell sequences by hippocampal cellular assemblies. Nature 469, 397–401 (2011).
McKenzie, S. et al. Preexisting hippocampal network dynamics constrain optogenetically induced place fields. Neuron 109, 1040–1054 (2021).
Valero, M., Zutshi, I., Yoon, E. & Buzsáki, G. Probing subthreshold dynamics of hippocampal neurons by pulsed optogenetics. Science 375, 570–574 (2022).
Villette, V., Malvache, A., Tressard, T., Dupuy, N. & Cossart, R. Internally recurring hippocampal sequences as a population template of spatiotemporal information. Neuron 88, 357–366 (2015).
Cavalieri, D. et al. CA1 pyramidal cell diversity is rooted in the time of neurogenesis. eLife 10, e69270 (2021).
Cossart, R. & Khazipov, R. How development sculpts hippocampal circuits and function. Physiol. Rev. 102, 343–378 (2022).
Angevine Jr, J. B. Time of neuron origin in the hippocampal region: an autoradiographic study in the mouse. Exp. Neurol. Suppl. 2, 1–70 (1965).
Deguchi, Y., Donato, F., Galimberti, I., Cabuy, E. & Caroni, P. Temporally matched subpopulations of selectively interconnected principal neurons in the hippocampus. Nat. Neurosci. 14, 495–504 (2011).
Druckmann, S. et al. Structured synaptic connectivity between hippocampal regions. Neuron 81, 629–640 (2014).
Lopes-dos-Santos, V., Ribeiro, S. & Tort, A. B. Detecting cell assemblies in large neuronal populations. J. Neurosci. Methods 220, 149–166 (2013).
Xu, H.-T. et al. Distinct lineage-dependent structural and functional organization of the hippocampus. Cell 157, 1552–1564 (2014).
Krumin, M. & Shoham, S. Generation of spike trains with controlled auto- and cross-correlation functions. Neural Comput. 21, 1642–1664 (2009).
Grosmark, A. D. & Buzsáki, G. Diversity in neural firing dynamics supports both rigid and learned hippocampal sequences. Science 351, 1440–1443 (2016).
English, D. F. et al. Pyramidal cell-interneuron circuit architecture and dynamics in hippocampal networks. Neuron 96, 505–520 (2017).
Harris, K. D., Csicsvari, J., Hirase, H., Dragoi, G. & Buzsáki, G. Organization of cell assemblies in the hippocampus. Nature 424, 552–556 (2003).
Lorente de N¢, R. Studies on the structure of the cerebral cortex. II. Continuation of the study of the ammonic system. J. Psychol. Neurol. 46, 113–177 (1934).
Hebb, D. O. The Organisation of Behaviour: A Neuropsychological Theory (Science Editions, 1949).
McClelland, J. L. Incorporating rapid neocortical learning of new schema-consistent information into complementary learning systems theory. J. Exp. Psychol. Gen. 142, 1190 (2013).
Hopfield, J. J. Neural networks and physical systems with emergent collective computational abilities. Proc. Natl Acad. Sci. USA 79, 2554–2558 (1982).
McKenzie, S. et al. Hippocampal representation of related and opposing memories develop within distinct, hierarchically organized neural schemas. Neuron 83, 202–215 (2014).
Tse, D. et al. Schemas and memory consolidation. Science 316, 76–82 (2007).
Edelman, G. M. Neural Darwinism: selection and reentrant signaling in higher brain function. Neuron 10, 115–125 (1993).
Battaglia, F. P., Sutherland, G. R., Cowen, S. L., Mc Naughton, B. L. & Harris, K. D. Firing rate modulation: a simple statistical view of memory trace reactivation. Neural Netw. 18, 1280–1291 (2005).
György Buzsáki, M. The Brain from Inside Out (Oxford Univ. Press, 2019).
Markram, H. et al. Interneurons of the neocortical inhibitory system. Nat. Rev. Neurosci. 5, 793–807 (2004).
Yoshimura, Y. & Callaway, E. M. Fine-scale specificity of cortical networks depends on inhibitory cell type and connectivity. Nat. Neurosci. 8, 1552–1559 (2005).
Li, Y. et al. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature 486, 118–121 (2012).
Ohtsuki, G. et al. Similarity of visual selectivity among clonally related neurons in visual cortex. Neuron 75, 65–72 (2012).
Farooq, U. & Dragoi, G. Emergence of preconfigured and plastic time-compressed sequences in early postnatal development. Science 363, 168–173 (2019).
Langston, R. F. et al. Development of the spatial representation system in the rat. Science 328, 1576–1580 (2010).
Dombeck, D. A., Harvey, C. D., Tian, L., Looger, L. L. & Tank, D. W. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci. 13, 1433–1440 (2010).
Noguchi, A., Huszár, R., Morikawa, S., Buzsáki, G. & Ikegaya, Y. Inhibition allocates spikes during hippocampal ripples. Nat. Commun. 13, 1280 (2022).
Rakic, P. in Evolutionary Developmental Biology of the Cerbral Cortex (eds Gregory R. Bock &, Gail Cardew) 30–45 (John Wiley, 2000).
Kitazawa, A. et al. Hippocampal pyramidal neurons switch from a multipolar migration mode to a novel ‘climbing’ migration mode during development. J. Neurosci. 34, 1115–1126 (2014).
Barabási, A.-L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).
Paxinos, G. & Franklin, K. B. Paxinos and Franklin’s The Mouse Brain in Stereotaxic Coordinates 5th edn (Academic Press, 2019).
Save, L., Baude, A. & Cossart, R. Temporal embryonic origin critically determines cellular physiology in the dentate gyrus. Cereb. Cortex 29, 2639–2652 (2019).
Dal Maschio, M. et al. High-performance and site-directed in utero electroporation by a triple-electrode probe. Nat. Commun. 3, 960 (2012).
Stancik, E. K., Navarro-Quiroga, I., Sellke, R. & Haydar, T. F. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J. Neurosci. 30, 7028–7036 (2010).
Donato, F., Jacobsen, R. I., Moser, M.-B. & Moser, E. I. Stellate cells drive maturation of the entorhinal-hippocampal circuit. Science 355, eaai8178 (2017).
Vöröslakos, M., Petersen, P. C., Vöröslakos, B. & Buzsáki, G. Metal microdrive and head cap system for silicon probe recovery in freely moving rodent. eLife 10, e65859 (2021).
Newman, J. P. et al. Optogenetic feedback control of neural activity. eLife 4, e07192 (2015).
Valero, M. et al. Sleep down state-active ID2/Nkx2. 1 interneurons in the neocortex. Nat. Neurosci. 24, 401–411 (2021).
Pachitariu, M., Steinmetz, N. A., Kadir, S. N., Carandini, M. & Harris, K. D. Fast and accurate spike sorting of high-channel count probes with KiloSort. Adv. Neural Inf. Process. Syst. 29, 4448–4456 (2016).
Petersen, P. C., Siegle, J. H., Steinmetz, N. A., Mahallati, S. & Buzs ki, G. CellExplorer: A framework for visualizing and characterizing single neurons. Neuron 109, 3594–3608 (2021).
Schmitzer-Torbert, N., Jackson, J., Henze, D., Harris, K. & Redish, A. Quantitative measures of cluster quality for use in extracellular recordings. Neuroscience 131, 1–11 (2005).
Levenstein, D., Buzsáki, G. & Rinzel, J. NREM sleep in the rodent neocortex and hippocampus reflects excitable dynamics. Nat. Commun. 10, 2478 (2019).
Tingley, D. & Buzsáki, G. Routing of hippocampal ripples to subcortical structures via the lateral septum. Neuron 105, 138–149 (2020).
Arenkiel, B. R. et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron 54, 205–218 (2007).
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005).
Lima, S. Q., Hromádka, T., Znamenskiy, P. & Zador, A. M. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS ONE 4, e6099 (2009).
Zhang, S.-J. et al. Optogenetic dissection of entorhinal-hippocampal functional connectivity. Science https://doi.org/10.1126/science.1232627 (2013).
Kvitsiani, D. et al. Distinct behavioural and network correlates of two interneuron types in prefrontal cortex. Nature 498, 363–366 (2013).
Stark, E. & Abeles, M. Unbiased estimation of precise temporal correlations between spike trains. J. Neurosci. Methods 179, 90–100 (2009).
Buzsáki, G. Theta oscillations in the hippocampus. Neuron 33, 325–340 (2002).
Peyrache, A., Khamassi, M., Benchenane, K., Wiener, S. I. & Battaglia, F. P. Replay of rule-learning related neural patterns in the prefrontal cortex during sleep. Nat. Neurosci. 12, 919–926 (2009).
van de Ven, G. M., Trouche, S., McNamara, C. G., Allen, K. & Dupret, D. Hippocampal offline reactivation consolidates recently formed cell assembly patterns during sharp wave-ripples. Neuron 92, 968–974 (2016).
Williams, C. K. & Rasmussen, C. E. Gaussian Processes for Machine Learning Vol. 2 (MIT Press, 2006).
Skaggs, W. E., McNaughton, B. L. & Gothard, K. M. in Advances in Neural Information Processing Systems 1030–1037 (Morgan Kauffman, 1993).
Buzsáki, G. & Mizuseki, K. The log-dynamic brain: how skewed distributions affect network operations. Nat. Rev. Neurosci. 15, 264–278 (2014).
Acknowledgements
We thank A. Fernandez-Ruiz, D. Huilgol, K. McClain, S. McKenzie, A. Navas-Olive, N. Nitzan, M. Valero and I. Zutshi for feedback on an early version of the manuscript. We also thank J. Dasen, Y. Fang, X. Wang, T. Yamaguchi and the Microscopy Laboratory at NYU Langone Health for technical support. This work was supported by grant no. U19NS104590-01and R01 MH122391 (to G.B.).
Author information
Authors and Affiliations
Contributions
R.H., H.B. and G.B. designed the study. R.H. and Y.Z. performed the experiments. R.H. performed data analysis and modeling with help from Y.Z. in analyzing histological data. R.H. and G.B. wrote the manuscript with contributions from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Roustem Khazipov and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
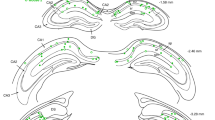
Extended Data Fig. 1 Birthdating CA1 pyramidal neurons via in utero electroporation.
a, Closeup (left) and full view (right) of an example coronal section shows the expression of ChR2-EYFP (green) and tdTomato (red) following in utero electroporation at E15.5. Expression was restricted to the hippocampal CA1 subregion in all n = 17 mice. b, Top: tdTomato expression in CA1 following in utero electroporation (different example from a). Manually drawn borders (dashed) delimit the pyramidal layer. The border between CA1 and CA2 (thick white line) was approximated by comparing with the Mouse Brain Atlas. White circles: tdTomato+ puncta identified via ImageJ. Bottom: Closeup of the projection foot of each tdTomato+ punctum on the border between stratum pyramidale (SP) and stratum radiatum (SR), from which each neuron’s radial depth was computed. Depth was normalized by the distance between the projection foot and crossing point with the border between SP and stratum oriens (SO). c, Top: Wideband activity recorded on a single silicon probe shank upon delivery of blue light pulses (2 ms; shaded blue). The bold channel highlights spikes (red dots) of a pyramidal neuron responding to blue light. Bottom: Autocorrelogram (left) of the highlighted neuron’s spike train reveals its burst firing, and the PSTH (middle) demonstrates its firing fidelity following light offset. Waveforms of spontaneous spikes are shown on the right (light blue: n = 50 spikes; dark blue: average). d, Left: Coronal section showing CA1 expression of tdTomato (red) introduced via in utero electroporation at E15.5. Cyan highlights nuclear expression of 5-bromo-2’deoxyuridine (BrdU), a marker of cell division and a commonly used birthdating tool. Right: A closeup highlighting three CA1 neurons exhibiting both tdTomato and BrdU expression. e, Fraction of Brdu and tdTomato double-labeled cells among tdTomato expressing neurons (mean ± SEM) as a function of BrdU injection timing (in hours) with respect to in utero electroporation at time 0. n = 3 animals per timepoint, >300 manually identified tdTomato+ neurons. The largest overlap occurred when BrdU was given at the time of in utero electroporation, whereas minimal overlap was seen when BrdU was injected 24 h after. This suggests that populations electroporated 24 h apart are distinct in terms of embryonic birthdate.
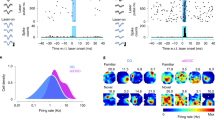
Extended Data Fig. 2 Clustering, classification and light responsiveness of CA1 neurons.
a, Filtered waveform trough-to-peak (ms) by burst index for isolated units (n = 9,471). Red: pyramidal neurons (n = 7,411). Black: narrow waveform interneurons (n = 1,556). Gray: wide waveform interneurons (n = 504). Blue crosses: light-responsive pyramidal neurons (n = 664, 8.96%). b, Left: Filtered waveforms (mean ± SD) of putative pyramidal neurons and interneurons. Right: Trough-to-peak bimodality used for neuron classification. c, Examples of light-responsive pyramidal neurons (as in Fig. 1d). d, Waveform profiles across recording channels for two neurons with maximum waveform amplitudes occurring on the same channel (‘nearby’ neurons, black triangle). Two sessions are shown. Blue: evoked spikes (<10 ms following light offset) of a light-responsive pyramidal neuron. Black: same neuron, n = 1000 spontaneous out-of-stimulation spikes. Red: non-responsive nearby pyramidal neuron. e, Waveforms in d, concatenated across channels. The L2 norm of vectorized waveform profile differences was used to quantify waveform profile distance. f, Left: Waveform profile distances between evoked and spontaneous spikes of light-responsive pyramidal neurons (blue, median=4.56; n = 664) and between evoked and spontaneous spikes of light-responsive and other nearby neurons, respectively (red, median=14.269; n = 3,086; P = 1.06e-255; two-sided Wilcoxon). Evoked spikes most resembled spontaneous spikes of their assigned cluster. Right: Waveform profile distances between SBD (blue, median=13.013; n = 1,743) and DBD pairs (red, median=15.68; n = 15,996; P = 4.6e-27; two-sided Wilcoxon) recorded on the same shank, irrespective of which channel. Only spontaneous spikes were considered. Additional analyses were performed to control for effects of waveform similarity in SBD populations (Extended Data Fig. 10). g, Top: PSTHs surrounding light pulse onset for statistically identified light-responsive pyramidal neurons (n = 664). Bottom: PSTH mean ± SEM of light-responsive (blue, n = 664) and non-responsive pyramidal neurons (n = 6,747). h, Cluster isolation quality (L-ratio)63 for light-responsive (blue, median=8.627; n = 664) and non-responsive pyramidal neurons (red, median=7.576; n = 6,747; P = 4.1e-3; two-sided Wilcoxon). Additional analyses were performed to control for effects of cluster isolation quality in SBD populations (Extended Data Fig. 5). i, Waveform shape trough-to-peak (ms) for light-responsive (blue, mean=0.696; n = 664) and non-responsive pyramidal neurons (red, mean=0.7; n = 6,747; P = 1.34e-1; two-sided, two-sample t-test). Inset: mean ± SD of filtered waveforms. Box plot's central mark, notch and edges indicate the median, its 95% confidence interval, and the 25th/75th percentiles, respectively.
Extended Data Fig. 3 Birthdate-dependent differences in single cell features cannot be explained by anatomical depth alone.
a, LFP profile around SPW-Rs. Sharp wave (SPW) slope estimated radial depth relative to the pyramidal layer center8,62. Neuron depth was given by the position of the channel where it exhibited maximal waveform amplitude (Extended Data Fig. 2d). b, Median depth of light-responsive pyramidal neurons (n = 664) occurred at −28.92 μm relative to SPW reversal. Deep and superficial (Sup) subpopulations were defined by a median-based split of the distribution. c, Left: Average firing rate. Deep, median=1.62 Hz (n = 332), Sup, median=1.39 Hz (n = 332); P = 3.1e-3, two-sided Wilcoxon. Middle: Burst index. Deep, median=0.119 (n = 332), Sup, median=0.123 (n = 332); P = 8.97e-1, two-sided Wilcoxon. Right: SPW-R participation probability. Deep, median=0.251 (n = 332), Sup, median=0.229 (n = 332); P = 4.8e-3, two-sided Wilcoxon. d, Left: Estimated depth. E13.5, −29 μm (n = 184 neurons); E14.5, −28.7 (n = 132); E15.5, −34.8 (n = 233); E16.5, −14.4 (115). Medians, Kruskal-Wallis: H = 26.327, df=3, P = 0. Middle: Neurons were resampled according to the distribution in b to match depth statistics while preserving n. Kruskal-Wallis: H = 3.65, df=3, P = 4.49e-1. Right: Burst indices (Fig. 1h) after matching depth distributions at preserved n. Kruskal-Wallis: H = 9.02, df=3, P = 5.2e-2. e, The procedure in d was repeated n = 500 times. Left: Light gray, median burst index across birthdates for each resampling. Black, average across resamplings. Right: P-value distribution (Kruskal-Wallis) across resamplings. Dashed vertical line, P = 0.05. 356/500 (71.2%) depth-matched burst index profiles fell below P = 0.05. Matching depth statistics weakens birthdate-dependent bursting differences, but preserves the bell-shaped profile, suggesting a residual effect of birthdate. f, Same as d-e, for SPW-R-related firing rates (Fig. 2f). 494/500 (98.8%) resamplings were P < 0.05. g, Same as d-e, for participation probability in SPW-Rs (Fig. 2f). 469/500 (93.8%) resamplings were P < 0.05. h, Same as d-e, for theta depth modulation (Fig. 2d). All resamplings were P < 0.05. i, Same as d-e, for spatial information scores (Fig. 5b). 363/500 (72.6%) resamplings were P < 0.05. j, Same as d-e, for spatial coverage (Fig. 5b). 369/500 (73.8%) resamplings were P = 0.05. Box plot central mark, notch and edges indicate the median, its 95% confidence interval, and the 25th/75th percentiles, respectively.
Extended Data Fig. 4 SPW-R correlations for pyramidal neuron pairs in individual mice.
Cumulative distributions and box plot summaries of pairwise correlations in SPW-Rs for pairs of SBD (blue) and DBD (red) neurons in individual animals. SBD pyramidal neurons exhibited higher cofiring in SPW-Rs than DBD pairs in 11/13 animals. Group sizes and P-values of two-sided Wilcoxon tests are shown above each box plot summary. Two animals electroporated at E13.5 were excluded due to a lack of SBD pairs. In these two mice only a single light-responsive pyramidal neuron per session was recorded. Box plot central mark, notch and edges indicate the median, its 95% confidence interval, and the 25th/75th percentiles, respectively.
Extended Data Fig. 5 Matching firing rate and L-ratio statistics supports results in Fig. 2c,g, Fig. 5c and Fig. 6e.
a, Left: Firing rate in SPW-Rs for SBD (blue, n = 3,751) and DBD (red, n = 50,461) pairs that contributed to pairwise correlation analyses in Fig. 2g. Right: Resampling the distribution of DBD pairs according to the empirical probability mass function of SBD pairs matches pairwise firing rate statistics and group sizes. b, Cofiring in SPW-Rs for SBD (blue, n = 3,751) and resampled DBD pairs (red, n = 3,751; P = 2.95e-15, two-sided Wilcoxon). c, Two-sided Wilcoxon P-values (n = 500 resamplings) for the comparison in b. All tests were P < 0.05. d, Same as a-c for theta cycle correlations (Fig. 2c) after n = 500 resamplings matching theta-related firing rates. All tests were P < 0.05. e, Same as a-c for spatial ratemap correlations (Fig. 5c) after n = 500 resamplings matching theta-related firing rates. 499/500 (99.8%) tests were P < 0.05. f, The same resampling as a matching L-ratio cluster isolation between SBD (blue, n = 3,751) and DBD (red, n = 50,461) pairs. g, Cofiring in SPW-Rs for SBD (blue, n = 3,751) and resampled DBD pairs (red, n = 3,751; P = 4.0001e-44, two-sided Wilcoxon). h, Two-sided Wilcoxon P-values (n = 500 resamplings) for the comparison in g. All P-values were < 0.05. i-j, Same as f-h for theta cycle (i) and spatial ratemap (j) correlations after n = 500 resamplings matching L-ratio statistics. All P-values were < 0.05. k, The same resampling as a matching theta-related firing rates of SBD (blue, n = 1,510) and DBD (red, n = 50,461) pairs that contributed to novel environment-related spatial ratemap correlations (Fig. 6e). l, Novel environment-related spatial ratemap correlations for SBD (blue, n = 1,510) and resampled DBD pairs (red, n = 1,510; P = 3.3e-6 two-sided Wilcoxon). m, Two-sided Wilcoxon P-values (n = 500 resamplings) for the comparison in l. 479/500 (95.8%) tests were P < 0.05. n-p, Same as k-m matching L-ratio cluster isolation statistics. p, 487/500 (97.4%) tests were P < 0.05. These results demonstrate that differences shown in Fig. 2c, g, Fig. 5c and Fig. 6e cannot be explained by firing rate, cluster isolation, or group size differences. Vertical dashed line indicates P = 0.05. Box plot central mark, notch and edges indicate the median, its 95% confidence interval, and the 25th/75th percentiles, respectively.
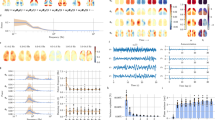
Extended Data Fig. 6 Birthdated pyramidal cells join assemblies comprising other SBD neurons.
a, Left: Raster plot illustrating conditional assembly detection during SPW-R-related spiking. SPW-Rs were binned at 25 ms resolution and concatenated. Assemblies were detected around spikes of a held-out, light-responsive pyramidal neuron (black triangle), with ICA performed on the remaining pyramidal neurons in time windows surrounding the held-out neuron’s spikes (vertical green bars). Right: Example independent components. Assembly members were determined as in Fig. 3. Assemblies with light-responsive members (blue, left) were compared to those consisting exclusively of non-responsive members (red, right). b, Probability of assembly expression around SPW-R-related spikes of the held-out neuron. Inset, Assembly expression strength at the time of the held-out neuron’s spike (blue, median=1.38, n = 1,109; red, median=1.235, n = 2,747; P = 2.29e-11; two-sided Wilcoxon). c, Pairwise spike count correlations in SPW-Rs between each held-out neuron and its SBD assembly members (blue, median=0.05, n = 1,153), DBD assembly members (red, median=0.0296, n = 15,906) and assembly non-members (gray, median=0.0242, n = 37,947). Kruskal-Wallis: H = 342.63, df=2, P = 0. d, Conditional assembly detection (as in a) during behavior on the familiar figure-eight maze (2.5 s binning). Right: Example independent components. e, Probability of assembly expression around spikes of the held-out neuron. Inset, Assembly expression strength at the time of the held-out neuron’s spike (blue, median=0.73, n = 1,705; red, median=0.663, n = 2,529; P = 9.9e-13; two-sided Wilcoxon). f, Spatial firing ratemap of a held-out, light-responsive pyramidal neuron (black), and assembly expression ratemaps of assemblies with light-responsive (dark and light blue) and non-responsive (red) members. Arrows, overlapping place fields. g, Correlations between held-out neurons’ ratemaps and assembly expression ratemaps for assemblies with light-responsive (blue, median=0.064; n = 1705) and non-responsive members (red, median=0.0057; n = 2529; P = 1.13e-08 two-sided Wilcoxon). h, Pairwise spike count correlations in theta cycles between each held-out neuron and its SBD assembly members (blue), DBD assembly members (red), and assembly non-members (gray). w/ SBD members, 0.0166 (n = 1611); w/ DBD members, 0.0046 (n = 18650); w/ non-members, 0.0023 (n = 25374). Medians, Kruskal-Wallis: H = 153.4885, df=2, P = 0. ***P < 0.001. Box plot central mark, notch and edges indicate the median, its 95% confidence interval, and the 25th/75th percentiles, respectively.
Extended Data Fig. 7 Expression of assemblies comprising exclusively SBD pyramidal neurons.
a, Left: Hard-coded weights define assemblies of different pyramidal neuron subgroups in an example session. This approach complements Fig. 3 where weights are assigned in an unsupervised manner via ICA. Blue: light-responsive neurons, black: non-responsive neurons. Top vector, assembly of light-responsive neurons (n = 27 members). Bottom two vectors, equal-sized assemblies with members randomly drawn from the non-responsive population (n = 147 in this session). Right: Time-resolved assembly expression during a SPW-R, obtained as in Fig. 3d. Significant peaks were taken as timepoints of assembly expression (Methods). b, Top: In an example session, SPW-R-related assembly expression rate (Hz) for an assembly consisting exclusively of light-responsive members (dashed blue) and for assemblies comprising randomly selected equal-sized subsets of non-responsive members (n = 100 groupings, gray histogram). The expression rate of the assembly comprising light-responsive members was expressed as a Z-score with respect to this distribution. Bottom: Z-scored expression rates of assemblies of light-responsive members across all sessions with ≥3 light-responsive pyramidal neurons. 62/65 (95.38%) values fell above zero, suggesting a greater propensity for higher order coactivation in assemblies of light-responsive members compared to equal-sized control assemblies. c, SPW-R-related firing of pyramidal neurons was binarized in each session. Instances of multiple spikes per SPW-R event were still recorded as ‘1’. For each SPW-R, the number of coactive light-responsive neurons (blue) was calculated. d, Probability of observing exactly k coactive (out of n = 11) light-responsive neurons (blue) in an example session. As control, random equal-sized subgroups of non-responsive neurons (n = 145 in this session) were selected, and the probability of k coactive neurons in SPW-Rs was computed. This was performed 1000 times (20 example traces are shown in gray), yielding a null distribution of probabilities for each k. Inset, Probability of k coactive light-responsive neurons expressed as a Z-score with respect to the null distribution. e, SPW-R-related probability of k-coactive light-responsive neurons (Z-scored; mean ± SEM) across all recording sessions with at least 2 light-responsive pyramidal neurons (n = 84 sessions). Light-responsive neurons of the same birthdate tend to coactivate in subgroups of more than 2 neurons.
Extended Data Fig. 8 Linear-nonlinear Poisson model for exploring assembly dynamics.
a, Schematic illustrating the linear-nonlinear Poisson model. A multivariate Gaussian distribution x with predefined mean and covariance was transformed using an exponential nonlinearity. The resulting lognormally distributed process λ was taken as the rate of a Poisson process to generate spike trains. kSE(dobdiff) is the kernel function for the Gaussian covariance, and depends on the difference of birthdates (dobdiff) between simulated neurons. b, Distribution of simulated birthdates of n = 350 neurons. Birthdates were sampled from a Gaussian with mean birthdate (E)14.5, and standard deviation of 1 day. c, Average firing rates of simulated neurons were set according to empirically observed firing rate distributions in SPW-Rs (Fig. 2f). <E14, n = 107; E14-E15, n = 68; E15-E16, n = 71; >E16, n = 104. d, Analytical covariance matrices (\(Cov\left( {{\uplambda}_i{\uplambda}_j} \right)\)) for two values of \(\sigma _{max}^2\), and a fixed value τ. These values were identical to the examples of simulated assembly expression rates in Fig. 4c. e, Simulated raster plot (top) and extracted assembly independent components (bottom) with kernel parameters \({\upsigma}_{max}^2 = 0.38\) and τ=4.44h. Arrows in the raster plot point to observable assembly expressions. Box plot central mark, notch and edges indicate the median, its 95% confidence interval, and the 25th/75th percentiles, respectively.
Extended Data Fig. 9 Uniform rate of neurogenesis fails to capture bell-shaped assembly dynamics.
a, Distribution of simulated birthdates of n = 350 neurons, sampled from a uniform distribution. b, Simulated assembly expression rates at different birthdates (blue dots) and their fit to assembly expression rates observed in data (red, average from Fig. 3e) depending on correlation strength \(\sigma _{max}^2\) between SBD neurons (same as Fig. 4c). c, Negative log probability (error) matrix quantifying goodness-of-fit of the uniform model (same color scale as Fig. 4d). White crosses show examples from b. d, Histogram of n = 100 error values obtained under the two parameter regimes highlighted in b. Each value was obtained from an independent sampling of the uniform model. Dashed vertical lines indicate the error under the bell-shaped model from Fig. 4. Single-tailed P-values were defined as the fraction of uniform model-generated error values that are smaller than the bell-shaped model-generated error for the same set of parameters. e, Matrix of thresholded P-values (as described in d) for the full range of model parameters. Magenta: P < 0.05; cyan: P > 0.05. The white line highlights a separation in the parameter space where the bell-shaped model performs no better than the uniform model (bottom left), and where the bell-shaped model outperforms the uniform (upper right).
Extended Data Fig. 10 Matching of waveform profile distances supports results in main Fig. 2c, g and Fig. 5c.
a, Left: Waveform profile distances (Extended Data Fig. 2d-f) for SBD (blue, median=13.0128, n=1,743) and DBD same-shank pairs (red, median=15.68, n=15,996; P=4.6e-27) contributing to analyses in Fig. 2c, g. Right: Waveform profile distances for same-shank pairs recorded >60 μm apart. Blue: median=17.41, n=347. Red, median=16.59, n=3,581. P=5.5e-1. Anatomical distance was defined as the distance between the neurons’ maximum waveform channels on the probe layout. Same-shank SBD and DBD pairs recorded >60 μm apart were less differentially affected by spike sorting errors, as evidenced by similar waveform profile distances. b, Left: SPW-R-related pairwise correlations (Fig. 2g) of same-shank SBD (blue, median=0.037, n=347) and DBD (red, median=0.021, n=3,588; P=1.4e-04) pairs recorded >60 μm apart. Right: Theta cycle correlations (Fig. 2c) for pairs shown on the left. Blue: SBD, median=0.019. Red: DBD, median=0.0092. P=4.3e-12. c, Left: Waveform profile distances of DBD pairs recorded >60 μm apart were resampled according to the empirical probability mass function of waveform profile distances of SBD pairs. Group size was fixed at n=1,743 of the original SBD group. >60 μm SBD pairs were sampled with replacement to achieve the latter. Blue: median=17.5854. Red: DBD, median=17.5305. P=6.9e-1. Right: P-value distribution (n=500 resamplings) for comparisons shown in b. All tests were P < 0.05. d, Same as a for same-shank pairs involved in spatial ratemap correlation analyses in Fig. 5c. Left: Blue: SBD, median=10.498, n=1,205. Red: DBD, median=14.49, n=13,549. P=6.5e-107. Right: Blue: SBD, median=14.0836, n=252. Red: DBD, median=15.028, n=2,998. P=7.8e-3. e, Spatial ratemap correlation of same-shank pairs recorded >60 μm apart. Blue: SBD, median=0.0079, n=252. Red: DBD, median=−0.0308, n=2998. P=3.3e-2. f, Same resampling as c matching waveform profile distances and group sizes of pairs involved in the spatial ratemap correlation analysis in Fig. 5c. Blue: SBD, median=14.0856. Red: DBD, median=13.97. P=3.1e-1. Right: P-value distribution (n=500 resamplings) for the comparison in e. 447/500 (89.4%) tests were P < 0.05. Vertical dashed line indicates P=0.05. Group comparisons were based on a two-sided Wilcoxon test. Box plot central mark, notch and edges indicate the median, its 95% confidence interval, and the 25th/75th percentiles, respectively.
Supplementary information
Supplementary Information
Supplementary Tables 1–5.
Rights and permissions
About this article
Cite this article
Huszár, R., Zhang, Y., Blockus, H. et al. Preconfigured dynamics in the hippocampus are guided by embryonic birthdate and rate of neurogenesis. Nat Neurosci 25, 1201–1212 (2022). https://doi.org/10.1038/s41593-022-01138-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01138-x
This article is cited by
-
The generative grammar of the brain: a critique of internally generated representations
Nature Reviews Neuroscience (2024)
-
Formation of memory assemblies through the DNA-sensing TLR9 pathway
Nature (2024)
-
Aversive stimulus-tuned responses in the CA1 of the dorsal hippocampus
Nature Communications (2023)
-
Backbone spiking sequence as a basis for preplay, replay, and default states in human cortex
Nature Communications (2023)