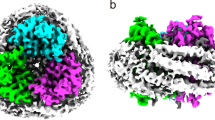
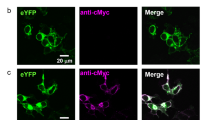
Abstract
Channelrhodopsins are used widely for optical control of neurons, in which they generate photoinduced proton, sodium or chloride influx. Potassium (K+) is central to neuron electrophysiology, yet no natural K+-selective light-gated channel has been identified. Here, we report kalium channelrhodopsins (KCRs) from Hyphochytrium catenoides. Previously known gated potassium channels are mainly ligand- or voltage-gated and share a conserved K+-selectivity filter. KCRs differ in that they are light-gated and have independently evolved an alternative K+ selectivity mechanism. The KCRs are potent, highly selective of K+ over Na+, and open in less than 1 ms following photoactivation. The permeability ratio PK/PNa of 23 makes H. catenoides KCR1 (HcKCR1) a powerful hyperpolarizing tool to suppress excitable cell firing upon illumination, demonstrated here in mouse cortical neurons. HcKCR1 enables optogenetic control of K+ gradients, which is promising for the study and potential treatment of potassium channelopathies such as epilepsy, Parkinson’s disease and long-QT syndrome and other cardiac arrhythmias.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The file ‘hyphochytrium_catenoides_predicted_proteins_renamed_modified.fasta’ that contains the sequence information of predicted H. catenoides proteins is available from https://www.ebi.ac.uk/biostudies/studies/S-BSST46. The whole genome shotgun sequences FLMG00000000.1 and CAFC00000000.2 are available from https://www.ncbi.nlm.nih.gov/nuccore/FLMG00000000.1 and https://www.ncbi.nlm.nih.gov/nuccore/CAFC00000000.2, respectively. The sequences of HcKCR1 and HcKCR2 expression constructs are available from GenBank (accession numbers MZ826861 and MZ826862, respectively). The plasmids encoding HcKCR1-mCherry, HcKCR2-mCherry, HcKCR1-EYFP and HcKCR2-EYFP in a mammalian expression vector backbone are available from Addgene (plasmids 177336, 177337, 182021 and 182022, respectively). Source data are provided with this paper.
Code availability
LogPro, custom software used for logarithmic filtration (noise reduction) of photocurrent traces, is freely available at Zenodo (https://zenodo.org/record/6461999#.Yl7Zx-jMIuF).
References
MacKinnon, R. Potassium channels. FEBS Lett. 555, 62–65 (2003).
Mironenko, A., Zachariae, U., de Groot, B. L. & Kopec, W. The persistent question of potassium channel permeation mechanisms. J. Mol. Biol. 433, 167002 (2021).
Sineshchekov, O. A., Jung, K.-H. & Spudich, J. L. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 99, 8689–8694 (2002).
Nagel, G. et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296, 2395–2398 (2002).
Nagel, G. et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl Acad. Sci. USA 100, 13940–13945 (2003).
Deisseroth, K. Optogenetics. Nat. Methods 8, 26–29 (2011).
Sineshchekov, O. A., Govorunova, E. G., Li, H. & Spudich, J. L. Bacteriorhodopsin-like channelrhodopsins: alternative mechanism for control of cation conductance. Proc. Natl Acad. Sci. USA 114, E9512–E9519 (2017).
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005).
Govorunova, E. G., Sineshchekov, O. A., Liu, X., Janz, R. & Spudich, J. L. Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650 (2015).
Mahn, M., Prigge, M., Ron, S., Levy, R. & Yizhar, O. Biophysical constraints of optogenetic inhibition at presynaptic terminals. Nat. Neurosci. 19, 554–556 (2016).
Messier, J. E., Chen, H., Cai, Z. L. & Xue, M. Targeting light-gated chloride channels to neuronal somatodendritic domain reduces their excitatory effect in the axon. eLife 7, e38506 (2018).
Richards, R. & Dempski, R. E. Re-introduction of transmembrane serine residues reduce the minimum pore diameter of channelrhodopsin-2. PLoS One 7, e50018 (2012).
Alberio, L. et al. A light-gated potassium channel for sustained neuronal inhibition. Nat. Methods 15, 969–976 (2018).
Beck, S. et al. Synthetic light-activated ion channels for optogenetic activation and inhibition. Front. Neurosci. 12, 643 (2018).
Bernal Sierra, Y. A. et al. Potassium channel-based optogenetic silencing. Nat. Commun. 9, 4611 (2018).
Leonard, G. et al. Comparative genomic analysis of the ‘pseudofungus’ Hyphochytrium catenoides. Open Biol. 8, 170184 (2018).
Tucker, K., Sridharan, S., Adesnik, H. & Brohawn, S. G. Cryo-EM structures of the channelrhodopsin ChRmine in lipid nanodiscs. Preprint at BioRxiv https://doi.org/10.1101/2021.11.21.469454 (2021).
Kishi, K. E. et al. Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Cell 185, 672–689.e623 (2022).
Hille, B. Ion Channels of Excitable Membranes (Sinauer Associates, 2001).
Eisenman, G. & R, H. Ionic selectivity revisited: the role of kinetic and equilibrium processes in ion permeation through channels. J. Membr. Biol. 76, 197–225 (1983).
Feldbauer, K. et al. Channelrhodopsin-2 is a leaky proton pump. Proc. Natl Acad. Sci. USA 106, 12317–12322 (2009).
Sineshchekov, O. A., Govorunova, E. G., Wang, J., Li, H. & Spudich, J. L. Intramolecular proton transfer in channelrhodopsins. Biophys. J. 104, 807–817 (2013).
Sineshchekov, O. A., Govorunova, E. G., Li, H. & Spudich, J. L. Gating mechanisms of a natural anion channelrhodopsin. Proc. Natl Acad. Sci. USA 112, 14236–14241 (2015).
Kuhne, J. et al. Unifying photocycle model for light adaptation and temporal evolution of cation conductance in channelrhodopsin-2. Proc. Natl Acad. Sci. USA 116, 9380–9389 (2019).
Ernst, O. P. et al. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 (2014).
Kandori, H. Biophysics of rhodopsins and optogenetics. Biophys. Rev. 12, 355–361 (2020).
Lanyi, J. K. Proton transfers in the bacteriorhodopsin photocycle. Biochim. Biophys. Acta 1757, 1012–1018 (2006).
Dreier, M. A. et al. Time-resolved spectroscopic and electrophysiological data reveal insights in the gating mechanism of anion channelrhodopsin. Commun. Biol. 4, 578 (2021).
Verhoefen, M. K. et al. The photocycle of channelrhodopsin-2: ultrafast reaction dynamics and subsequent reaction steps. ChemPhysChem 11, 3113–3122 (2010).
Govorunova, E. G., Sineshchekov, O. A. & Spudich, J. L. Structurally distinct cation channelrhodopsins from cryptophyte algae. Biophys. J. 110, 2302–2304 (2016).
Yamauchi, Y. et al. Molecular properties of a DTD channelrhodopsin from Guillardia theta. Biophys. Physicobiol. 14, 57–66 (2017).
Sineshchekov, O. A. et al. Conductance mechanisms of rapidly desensitizing cation channelrhodopsins from cryptophyte algae. mBio 11, e00657–00620 (2020).
Oppermann, J. et al. MerMAIDs: a family of metagenomically discovered marine anion-conducting and intensely desensitizing channelrhodopsins. Nat. Commun. 10, 3315 (2019).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Govorunova, E. G. et al. RubyACRs, non-algal anion channelrhodopsins with highly red-shifted absorption. Proc. Natl Acad. Sci. USA 117, 22833–22840 (2020).
Hoffmann, M. et al. Color tuning in rhodopsins: the mechanism for the spectral shift between bacteriorhodopsin and sensory rhodopsin II. J. Am. Chem. Soc. 128, 10808–10818 (2006).
Wiegert, J. S., Mahn, M., Prigge, M., Printz, Y. & Yizhar, O. Silencing neurons: tools, applications, and experimental constraints. Neuron 95, 504–529 (2017).
Plugge, B. et al. A potassium channel protein encoded by chlorella virus PBCV-1. Science 287, 1641–1644 (2000).
Prakash, R. et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat. Methods 9, 1171–1179 (2012).
Adesnik, H. & Abdeladim, L. Probing neural codes with two-photon holographic optogenetics. Nat. Neurosci. 24, 1356–1366 (2021).
Papagiakoumou, E. et al. Scanless two-photon excitation of channelrhodopsin-2. Nat. Methods 7, 848–854 (2010).
Govorunova, E. G. et al. Cation and anion channelrhodopsins: sequence motifs and taxonomic distribution. MBio 12, e0165621 (2021).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Spudich, J. L. LogPro. Zenodo https://doi.org/10.5281/zenodo.6461999 (2022).
Waschuk, S. A., Bezerra, A. G. J., Shi, L. & Brown, L. S. Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eukaryote. Proc. Natl Acad. Sci. USA 102, 6879–6883 (2005).
Xue, M., Atallah, B. V. & Scanziani, M. Equalizing excitation-inhibition ratios across visual cortical neurons. Nature 511, 596–600 (2014).
Acknowledgements
This work was supported by the National Institutes of Health grants R35GM140838 (J.L.S.), U01NS118288 (M.X., J.L.S. and F.St.-P.), R01EB027145 (F.St.-P.), U01NS113294 (F.St.-P.), and P50HD103555 (Baylor College of Medicine Intellectual and Developmental Disabilities Research Center, Neurovisualization Core); National Science Foundation grants 1707359 and 1935265 (F.St.-P.); Robert A. Welch Foundation Endowed Chair AU-0009 (J.L.S.); Welch Foundation grant Q-2016-20190330 (F.St.-P.); McKnight Endowment Fund for Neuroscience (M.X.); Natural Sciences and Engineering Research Council of Canada Discovery Grant RGPIN-2018-04397 (L.S.B.); a Klingenstein-Simons Fellowship Award in Neuroscience (F.St.-P.). M.X. is a Caroline DeLuca Scholar. F.St.-P. is a Scholar of the McNair Medical Foundation. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
E.G.G., O.A.S., L.S.B., M.X., F.St.-P. and J.L.S. conceptualized the work and developed its methodology. L.S.B. identified and refined the HcKCR sequences in the public databases. E.G.G. and O.A.S. carried out patch clamp experiments in HEK293 cells, analyzed their results and prepared the corresponding figures. H.L. and Y.W. expressed and purified HcKCR1 from Pichia. O.A.S. carried out flash photolysis experiments, analyzed their results and prepared the corresponding figures. Y.G. carried out neuronal experiments under supervision of M.X., and they both analyzed the results and prepared the corresponding figures. X.L. designed, conducted, and analyzed the 2P photoactivation experiments under the supervision of F.St.-P. X.L. and F.St.-P. prepared the corresponding figures. L.S.B., M.X., F.St.-P. and J.L.S. provided the funds. M.X. and J.L.S. supervised and administered the project. E.G.G., O.A.S., Y.G., X.L., M.X., F.St.-P. and J.L.S. wrote an original draft, and all authors contributed to its review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Anna Morini, Ofer Yizhar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The alignment of KCRs and representative cryptophyte BCCRs.
The black lines show the predicted transmembrane helices (TM1-TM7). The Schiff base lysine is highlighted blue, the conserved aspartates corresponding to Asp85 and Asp96 of bacteriorhodopsin, red, and the residues implicated in trimer formation in ChRmine, green. GtCCR2, Guillardia theta CCR2; RaCCR1, Rhodomonas abbreviata CCR1.
Extended Data Fig. 2 Electrophysiological characterization of HcKCR2 in HEK293 cells.
a, b, Series of HcKCR2 photocurrents recorded in response to 1-s light pulses under indicated ionic and voltage conditions. c, The IV curves measured under indicated ionic conditions (mean ± sem, n = 7 cells from 3 independent experiments). d, e, The Vrev values determined for the peak current and current at the end of a 1-s light pulse under indicated ionic conditions. The symbols are data from individual cells, the lines are the mean ± sem, n = 7 cells from 3 independent experiments. *, P = 0.031; n. s. (not significant), P = 1 by two-sided paired sample Wilcoxon signed ranks test. Source data are provided.
Extended Data Fig. 3 Analysis of relative permeabilities of HcKCR1 for metal cations and NMDG+.
Left, HcKCR1 peak photocurrents recorded in response to 1-s light pulses with 130 mM KCl in the pipette and 130 mM of the indicated cation in the bath. Right, the corresponding IV curves (mean ± sem, n = 8 cells from 3 independent experiments). The red lines show linear approximations used to determine the Vrev. Source data are provided.
Extended Data Fig. 4 Comparison of the Vrev values of HcKCR1 photocurrents measured at the time of the peak and at the end of a 1-s light pulse.
The photocurrents were recorded with 130 mM KCl in the pipette and 130 mM of the indicated cation in the bath. The symbols are the data from individual cells, the lines are the mean ± sem, n = 10 cells for Na+, 7 cells for Li+, Cs+, NMDG+, Ca2+, K+ pH 5.4, 6 cells for Mg2+, and 8 cells for K+ pH 7.4 and Rb+. P values were determined by two-sided paired sample Wilcoxon signed ranks test. Source data are provided.
Extended Data Fig. 5 Light intensity dependence of HcKCR1 photocurrents.
a, A series of HcKCR1 photocurrents recorded at 20 mV in response to 200-ms light pulses of the intensity indicated in the legend. The green bar shows the duration of illumination. b–f, Dependence of peak photocurrent (b), photocurrent at the end of the light pulse (c), desensitization (d), the time constant (τ) of photocurrent rise (e), and τ of photocurrent decay (f) on the stimulus intensity. Light gray, data from individual cells, and black, mean ± sem (n = 5 cells from 3 independent experiments). Source data are provided.
Extended Data Fig. 6 Representative photocurrents recorded under two-photon (2P) illumination at different laser powers and wavelengths.
a, b, Representative HcKCR1 (a) and HcKCR2 (b) photocurrents recorded at different power levels. c, d, Representative HcKCR1 (c) and HcKCR2 (d) photocurrents recorded with light of different wavelengths. All traces were recorded using KCR-expressing HEK293A cells held at −20 mV. The red bars mark the timing of the 1-s 2P excitation periods. The jagged appearance of the traces is due to raster scanning artifacts (see Methods).
Extended Data Fig. 7 Extended analysis of photocurrents and photochemical conversion upon single quantum excitation.
a, Photocurrent traces (thin solid lines) recorded from HcKCR1 in response to 6-ns laser flashes at 20-mV voltage increments under indicated ionic conditions and their multiexponential approximations (dashed lines). b, The time constants (τ) of the three kinetic components of channel currents at −60 (red) and 0 (black) mV (mean ± sem). The data for individual cells are shown as circles; mean ± sem, as lines (n = 16 cells for fast opening, 13 cells for slow opening at −60 mV, 10 cells for slow opening at 0 mV, and 17 cells for closing). *, P = 0.017 **, P = 0.06; n.s. (not significant), P = 0.877 by the two-sided Mann–Whitney test. Statistics source data are provided. c, The voltage dependence of the three kinetic components of channel currents. d, Peak amplitude channel currents recorded at varied time intervals between laser flashes. The datapoints are mean ± sem, n = 5 cells. Statistics source data are provided. e, Laser flash-induced absorption changes of HcKCR1 in detergent (black) and Pichia membranes (red). Experimental data are shown as thin solid lines, and their multiexponential approximations, as dashed lines. f, HcKCR1 photocurrent traces in the absence of permeant metal cations at bath pH 5.4. Experimental data are shown as thin solid lines, and their multiexponential approximations, as dashed lines.
Extended Data Fig. 8 HcKCR1 expression in mouse cortical neurons.
a, Fluorescence images showing HcKCR1-EYFP (green) and tdTomato (magenta) expression in layer 2/3 pyramidal neurons. HcKCR1-EYFP is expressed at high levels and forms some intracellular aggregates, as do many other wild-type ChRs. HcKCR2-EYFP shows the same degree of aggregation. Membrane targeting of both KCRs is confirmed by robust photocurrents (Fig. 5b and Extended Data Fig. 9). b, The fluorescence image of HcKCR1-EYFP from Fig. 5a (left panel) was overexposed to visualize the presence of HcKCR1-EYFP in the dendrites and axons (right panel). Note, the axons of layer 2/3 pyramidal neurons ramify in layer 5. Similar results were observed in 14 slices from 2 male and 2 female mice at the age of 3–4 weeks.
Extended Data Fig. 9 Photoactivation of HcKCR2 in neurons causes action potentials, but can also inhibit neuronal spiking.
a, Photocurrent traces of a HcKCR2-expressing neuron in response to a 1-s 470 nm light pulse (18.0 mW mm-2) at holding voltages increased in 5-mV steps. HcKCR2 generated robust photocurrents, but at more negative voltages the onset of illumination caused large inward currents, corresponding to action potentials (boxed segment, see its expansion in b). b, Expanded photocurrent traces of the boxed segment in a. c, d, IV curves of the early photocurrent before the onset of action potentials (c) and the current at the end of illumination (d) of individual neurons indicated by different colors (P < 0.0001, R2 > 0.99 for all linear regressions). e, Reversal potentials calculated from the data in c and d (P = 0.1, two-tailed paired t test). The reversal potentials of HcKCR2 are higher than those of HcKCR1, consistent with the results in HEK293 cells (Extended Data Fig. 2). f, Membrane voltage traces of a HcKCR2-expressing neuron in response to −0.1 (left), 0 (middle), or 0.5 nA (right) current injections without (top) and with (bottom) 470 nm illumination. When the membrane potential was at rest or depolarized (middle and right), photoactivation of HcKCR2 often caused action potentials at the onset of light because the reversal potential was close to the action potential threshold. Nevertheless, action potentials evoked by current injections were suppressed by long pulses of light (right) due to shunting inhibition. g, The frequencies of action potentials evoked by different current injections with (magenta) and without (black) photoactivation (P > 0.99 for all current levels by the Multiple Wilcoxon matched-pairs signed rank test with Bonferroni–Dunn multiple corrections). Data in e and g are expressed as mean ± sem, n = 3 neurons from 1 male and 1 female mouse at the age of 3–4 weeks.
Extended Data Fig. 10 HcKCR1 photocurrent recovery in the dark in experiments with 1-s light pulses.
Neurons were stimulated by two light pulses (13.1 mW mm−2) applied with a 10-s, 20-s, or 30-s interval, and the ratio of the peak or end currents evoked by the 2nd pulse to that of the 1st pulse was calculated. ITI, intertrial interval. Data are expressed as mean ± sem; n = 8 neurons from 1 male and 1 female mouse at the age of 3–4 weeks.
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Govorunova, E.G., Gou, Y., Sineshchekov, O.A. et al. Kalium channelrhodopsins are natural light-gated potassium channels that mediate optogenetic inhibition. Nat Neurosci 25, 967–974 (2022). https://doi.org/10.1038/s41593-022-01094-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01094-6
This article is cited by
-
Kalium channelrhodopsins effectively inhibit neurons
Nature Communications (2024)
-
Structures of channelrhodopsin paralogs in peptidiscs explain their contrasting K+ and Na+ selectivities
Nature Communications (2023)
-
A blue-shifted anion channelrhodopsin from the Colpodellida alga Vitrella brassicaformis
Scientific Reports (2023)
-
Cardiac optogenetics: shining light on signaling pathways
Pflügers Archiv - European Journal of Physiology (2023)
-
Optogenetics meets physiology
Pflügers Archiv - European Journal of Physiology (2023)