Abstract
Excitatory neurons are preferentially impaired in early Alzheimer’s disease but the pathways contributing to their relative vulnerability remain largely unknown. Here we report that pathological tau accumulation takes place predominantly in excitatory neurons compared to inhibitory neurons, not only in the entorhinal cortex, a brain region affected in early Alzheimer’s disease, but also in areas affected later by the disease. By analyzing RNA transcripts from single-nucleus RNA datasets, we identified a specific tau homeostasis signature of genes differentially expressed in excitatory compared to inhibitory neurons. One of the genes, BCL2-associated athanogene 3 (BAG3), a facilitator of autophagy, was identified as a hub, or master regulator, gene. We verified that reducing BAG3 levels in primary neurons exacerbated pathological tau accumulation, whereas BAG3 overexpression attenuated it. These results define a tau homeostasis signature that underlies the cellular and regional vulnerability of excitatory neurons to tau pathology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data used to generate the results that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Šimić, G. et al. Tau protein hyperphosphorylation and aggregation in Alzheimer’s disease and other tauopathies, and possible neuroprotective strategies. Biomolecules 6, 6 (2016).
Dugger, B. N. & Dickson, D. W. Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 9, a028035 (2017).
Spina, S. et al. The tauopathy associated with mutation+3 in intron 10 of Tau: characterization of the MSTD family. Brain 131, 72–89 (2008).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Davies, P. & Maloney, A. J. Selective loss of central cholinergic neurons in Alzheimer’s disease. Lancet 2, 1403 (1976).
Morrison, B. M., Hof, P. R. & Morrison, J. H. Determinants of neuronal vulnerability in neurodegenerative diseases. Ann. Neurol. 44(Suppl 1), S32–S44 (1998).
Götz, J., Schonrock, N., Vissel, B. & Ittner, L. M. Alzheimer’s disease selective vulnerability and modeling in transgenic mice. J. Alzheimers Dis. 18, 243–251 (2009).
Riascos, D. et al. Age-related loss of calcium buffering and selective neuronal vulnerability in Alzheimer’s disease. Acta Neuropathol. 122, 565–576 (2011).
Ahmadian, S. S. et al. Loss of calbindin-D28K is associated with the full range of tangle pathology within basal forebrain cholinergic neurons in Alzheimer’s disease. Neurobiol. Aging 36, 3163–3170 (2015).
Hof, P. R. et al. Age-related distribution of neuropathologic changes in the cerebral cortex of patients with Down’s syndrome. Quantitative regional analysis and comparison with Alzheimer’s disease. Arch. Neurol. 52, 379–391 (1995).
Seeley, W. W. Selective functional, regional, and neuronal vulnerability in frontotemporal dementia. Curr. Opin. Neurol. 21, 701–707 (2008).
Hyman, B. T., Van Hoesen, G. W., Damasio, A. R. & Barnes, C. L. Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science 225, 1168–1170 (1984).
Mattson, M. P. & Magnus, T. Ageing and neuronal vulnerability. Nat. Rev. Neurosci. 7, 278–294 (2006).
Saxena, S. & Caroni, P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron 71, 35–48 (2011).
Roselli, F. & Caroni, P. From intrinsic firing properties to selective neuronal vulnerability in neurodegenerative diseases. Neuron 85, 901–910 (2015).
Labbadia, J. & Morimoto, R. I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 (2015).
Taylor, R. C. & Dillin, A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3, a004440 (2011).
Kaushik, S. & Cuervo, A. M. Proteostasis and aging. Nat. Med. 21, 1406–1415 (2015).
Freer, R. et al. A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer’s disease. Sci. Adv. 2, e1600947 (2016).
Liu, L. et al. Trans-synaptic spread of tau pathology in vivo. PLoS One 7, e31302 (2012).
Lei, Z., Brizzee, C. & Johnson, G. V. BAG3 facilitates the clearance of endogenous tau in primary neurons. Neurobiol. Aging 36, 241–248 (2015).
Seidel, K. et al. The HSPB8-BAG3 chaperone complex is upregulated in astrocytes in the human brain affected by protein aggregation diseases. Neuropathol. Appl. Neurobiol. 38, 39–53 (2012).
Wegmann, S. et al. Removing endogenous tau does not prevent tau propagation yet reduces its neurotoxicity. EMBO J. 34, 3028–3041 (2015).
Lake, B. B. et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-Seq. Nat. Methods 14, 955–958 (2017).
Ciryam, P. et al. A transcriptional signature of Alzheimer’s disease is associated with a metastable subproteome at risk for aggregation. Proc. Natl Acad. Sci. USA 113, 4753–4758 (2016).
Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, e17 (2005).
Fu, H., Hardy, J. & Duff, K. E. Selective vulnerability in neurodegenerative diseases. Nat. Neurosci. 21, 1350–1358 (2018).
de Calignon, A. et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 73, 685–697 (2012).
Fu, H. et al. 3D visualization of the temporal and spatial spread of tau pathology reveals extensive sites of tau accumulation associated with neuronal loss and recognition memory deficit in aged tau transgenic mice. PLoS One 11, e0159463 (2016).
Ferrer, I. et al. Glial and neuronal tau pathology in tauopathies: characterization of disease-specific phenotypes and tau pathology progression. J. Neuropathol. Exp. Neurol. 73, 81–97 (2014).
Kahlson, M. A. & Colodner, K. J. Glial tau pathology in tauopathies: functional consequences. J. Exp. Neurosci. 9(Suppl 2), 43–50 (2016).
Leyns, C. E. G. & Holtzman, D. M. Glial contributions to neurodegeneration in tauopathies. Mol. Neurodegener. 12, 50 (2017).
Brehme, M. et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 9, 1135–1150 (2014).
Baranczak, A. & Kelly, J. W. A current pharmacologic agent versus the promise of next generation therapeutics to ameliorate protein misfolding and/or aggregation diseases. Curr. Opin. Chem. Biol. 32, 10–21 (2016).
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008).
Seubert, P. et al. Detection of phosphorylated Ser262 in fetal tau, adult tau, and paired helical filament tau. J. Biol. Chem. 270, 18917–18922 (1995).
Sanders, D. W. et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288 (2014).
Fu, H. et al. Tau pathology induces excitatory neuron loss, grid cell dysfunction, and spatial memory deficits reminiscent of early Alzheimer’s disease. Neuron 93, 533–541.e5 (2017).
Tasic, B. et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346 (2016).
Ji, C., Steimle, B. L., Bailey, D. K. & Kosman, D. J. The ferroxidase hephaestin but not amyloid precursor protein is required for ferroportin-supported iron efflux in primary hippocampal neurons. Cell. Mol. Neurobiol. 38, 941–954 (2018).
Tosto, G. et al. F-box/LRR-repeat protein 7 is genetically associated with Alzheimer’s disease. Ann. Clin. Transl. Neurol 2, 810–820 (2015).
Wu, J. W. et al. Small misfolded Tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. J. Biol. Chem. 288, 1856–1870 (2013).
Acknowledgements
We thank M. Diamond (UT Southwestern Medical Center) for providing the Tau RD-P301S-YFP lentivirus and DS9 clone cell line; K. Zhang and B. Lake (UCSD) and A. Regev, N. Habib, I. Avraham-Davidi, and A. Basu (Broad Institute of MIT and Harvard) for sharing their single-nucleus RNA-seq datasets; L. Honig, J.P. Vonsattel, A. Teich, and E. Cortés (New York Brain Bank, Columbia University Medical Center), the NIH NeuroBrainBank at the University of Maryland Brain and Tissue Bank as well as T. Beach and G. Serrano (Banner Sun Health Research Institute Brain and Body Donation Program) for providing human de-identified brain tissue and for helpful discussions on brain regions and immunostaining techniques; P. Davies (The Feinstein Institute for Medical Research) for providing the MC1 and PHF1 tau antibodies; P. Dolan (Prothena) for providing the 12E8 tau antibody; C. Pröschel (University of Rochester Medical Center) for providing psPAX2 and VSVG; ACDBio for troubleshooting on RNAscope FISH; and W.H. Yu and C.L. Clelland for discussing the results. This work was funded by: NIH/NIA AG056673 (H.F.), Alzheimer’s Association AARF-17-505009 (H.F.), NIH/NINDS NS074874 (K.E.D.), NIH/NIA AG056151 (K.E.D) and by the BrightFocus Foundation, the Rainwater Foundation/Tau Consortium and the Cure Alzheimer’s Fund (K.E.D). The Banner Sun Health Research Institute Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium), and the Michael J. Fox Foundation for Parkinson’s Research.
Author information
Authors and Affiliations
Contributions
K.E.D., H.F., and M.V. jointly designed and supervised the study, discussed the results, and wrote the paper. H.F. designed and performed experiments and analyzed the data. A.P., R.F., and M.V. analyzed single-nucleus RNA-seq datasets. A.P. discussed the results and wrote parts of the paper. Y.N., N.C.H.V., M.T., P.V.M.C., B.A.L., S.L.F., S.C., and H.Y.F. provided technical assistance. E.D.H. provided critical input into the paper. G.V.W.J. designed and supplied all the BAG3-related viruses. All authors discussed the results and contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
K.E.D. is on the board of directors and SAB of Ceracuity LLC. All other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Tau pathology co-localizes with EX, but not with IN neurons in secondary affected regions in EC-tau mice.
(a-d) Representative images of MC1+ tau staining co-localized with TBR1+ and SATB2+ EX neurons, but not PVALB+, SST+ or CALB2+ IN neurons, in the PRH (a) and NC (b) of EC-tau mice at 22 months (n = 6 animals, 2 sections each animal), and in the PRH (c) and NC (d) of EC-tau mice at 30+ months (n = 5-6 animals, 2 sections each animal). Scale bar, 20 µm.
Supplementary Figure 2 Tau pathology co-localizes with EX, but not with IN neurons or glial cells in human AD brain.
(a) Representative images of MC1+ tau staining co-localized with TBR1+ and SATB2+ EX neurons, but not PVALB+, SST+ or CALB2+ IN neurons in the prefrontal cortex (BA9) of human AD brain at Braak stage V-VI. (b, c) Representative images of AT8+ tau staining did not co-localize with IBA-1+ microglia (b) or GFAP+ astrocytes (c) in the EC of human AD brain at different Braak stages. Scale bars, 20 µm (a) and 10 µm (b and c). Three independent experiments were repeated with similar results.
Supplementary Figure 3 Tau pathology detected by specific phospho-tau antibodies co-localizes with EX, but not IN, neurons in EC-tau mice and in human AD brain.
(a) Representative images of phospho-tau staining (AT8+, PHF1+, pS422+) co-localized with SATB2+ EX neurons, but not GAD1+ IN neurons in the MEC of EC-Tau mice at 22 months. (b) Representative images of phospho-tau staining (AT8+, PHF1+, AT100+) co-localized with SATB2+ EX neurons, but not GAD1+ IN neurons in the BA9 of human AD at Braak stage V-VI. Scale bar, 20 µm. Three independent experiments were repeated with similar results.
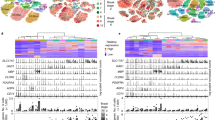
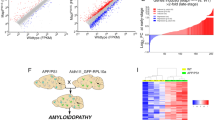
Supplementary Figure 4 Null model for seven different subproteomes between EX and IN neurons.
The random distribution of the mean values (orange histogram) computed on each sample, the normal fit (solid orange line), and the observed value (dashed red vertical line) are reported in each panel (see Methods for details). Next to each observed value, the probability computed with the cumulative distribution obtained from the fit and evaluated at the observed mean is reported. (a-g) Null models from the SNS dataset; (h-n) Null models from the DroNc-Seq dataset. Here nsns and ndrnc represent the sample sizes corresponding to SNS and DroNc-seq datasets, respectively. (a, h) EX markers:): nsns=ndrnc=2; (b, i) tau: nsns=ndrnc=1; c, j) promoters (tau aggregation promoters): nsns=ndrnc=6; (d, k) MS (metastable subproteome): nsns=162, ndrnc=179; (e, l) tangles (tangle co-aggregators): nsns=57, ndrnc=68; f, m) protectors (tau aggregation protectors): nsns=ndrnc=6; (g, n) IN markers: nsns=ndrnc=3. A p-value is computed as the probability to have a value more extreme than the observed one (one-tailed).
Supplementary Figure 5 Null model for five different subproteomes in EX neurons between early- and late-affected brain regions.
The statistical significance of the results is studied by creating a null model for each subproteome under scrutiny (see Supplementary Figure 4). (a-e) Null models from the SNS dataset (BA21+22+10+41 vs BA17); (f-j) Null models from the DroNc-Seq dataset (HP vs PFC). (a, f) protectors; (b, g) promoters; (c, h) MS; (d, i) tangles; (e, j) tau. A p-value is computed as the probability to have a value more extreme than the observed one (one-tailed). Sample sizes are the same as Supplementary Figure 4.
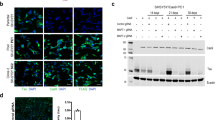
Supplementary Figure 7 BAG3 protein is differentially regulated in neurons in human non-AD and AD cases.
(a) Representative immunofluorescent images of the co-staining of IN neuronal marker (GAD1, purple), pan neuronal marker (NeuN, green), and BAG3 (red) in the BA9 of human AD (Braak stage V/VI) brain. The white arrow represents the IN neurons (GAD1+/NeuN+), while the white arrowhead indicates the putative EX neurons (GAD1-/NeuN+). The letter “g” stands for glial cells. The nuclei (blue) were counterstained with Hoechest3342. Three independent experiments were repeated with similar results. Scale bar, 20 µm. (b) Comparison of the mean intensity of BAG3 in individual neurons in the BA9 regions (n = 3 human brains, 20 GAD1-/NeuN+ and 20 GAD1+/NeuN+ neurons from each case). Data are presented as mean ± SEM. The statistical significance was assessed by the two-tailed nonparametric Mann-Whitney test. *** P < 0.0001 vs non-AD and/or AD EX neurons (The Mann-Whitney U is 1551, 714, 531, and 130, respectively).
Supplementary Figure 8 Null model for seven different subproteomes between microglia and EX or IN neurons.
The statistical significance of the results is studied by creating a null model for each subproteome under scrutiny (see Supplementary Figure 4). (a, c, e, g) Null models for MG (microglia) vs EX (excitatory) neurons; (b, d, f, h) Null models for MG (microglia) vs IN (inhibitory) neurons. (a, b) tau; (c, d) promoters; (e, f) tangles; (g, h) protectors. A p-value is computed as the probability to have a value more extreme than the observed one (one-tailed). Sample sizes are the same as Supplementary Figure 4.
Supplementary Figure 9 Null model for seven different subproteomes between astrocytes and EX or IN neurons.
The statistical significance of the results is studied by creating a null model for each subproteome under scrutiny (see Supplementary Figure 4). (a, c, e, g) Null models for ASC (astrocytes) vs EX (excitatory) neurons; (b, d, f, h) Null models for ASC (astrocytes) vs IN (inhibitory) neurons. (a, b) tau; (c, d) promoters; (e, f) tangles; (g, h) protectors. A p-value is computed as the probability to have a value more extreme than the observed one (one-tailed). Sample sizes are the same as Supplementary Figure 4.
Supplementary Figure 10 Null model for seven different subproteomes between oligodendrocytes and EX or IN neurons.
The statistical significance of the results is studied by creating a null model for each subproteome under scrutiny (see Supplementary Figure 4). (a, c, e, g) Null models for ODC (oligodendrocytes) vs EX (excitatory) neurons; (b, d, f, h) Null models for ODC (oligodendrocytes) vs IN (inhibitory) neurons. (a, b) tau; (c, d) promoters; (e, f) tangles; (g, h) protectors. A p-value is computed as the probability to have a value more extreme than the observed one (one-tailed). Sample sizes are the same as Supplementary Figure 4.
Supplementary Figure 11 Full length of the western blot image.
Original western blot images of Fig. 7a in primary cortical neurons transduced with lentivirus expressing scrambled BAG3 or shBAG3, or overexpressing BAG3 (OE) as described in Methods. BAG3 is the target protein detected by primary rabbit anti-BAG3 antibody. GAPDH is a housekeeping protein used as the loading control.
Supplementary information
Supplementary Figures 1–11
Supplementary Figures 1–11
Supplementary Table 1
Single-nucleus RNA-seq datasets analysis in neurons.
Supplementary Table 2
Single-nucleus RNA-seq datasets analysis in glia.
Supplementary Table 3
Weighted gene co-expression network analysis of the subproteomes relevant to tau homeostasis.
Supplementary Table 4
Supplementary Table 4.
Rights and permissions
About this article
Cite this article
Fu, H., Possenti, A., Freer, R. et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat Neurosci 22, 47–56 (2019). https://doi.org/10.1038/s41593-018-0298-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-018-0298-7
This article is cited by
-
Molecular and cellular mechanisms of selective vulnerability in neurodegenerative diseases
Nature Reviews Neuroscience (2024)
-
DLK-MAPK Signaling Coupled with DNA Damage Promotes Intrinsic Neurotoxicity Associated with Non-Mutated Tau
Molecular Neurobiology (2024)
-
Targeting vulnerable microcircuits in the ventral hippocampus of male transgenic mice to rescue Alzheimer-like social memory loss
Military Medical Research (2024)
-
Whole-brain modeling of the differential influences of amyloid-beta and tau in Alzheimer’s disease
Alzheimer's Research & Therapy (2023)
-
Proteostasis failure exacerbates neuronal circuit dysfunction and sleep impairments in Alzheimer’s disease
Molecular Neurodegeneration (2023)