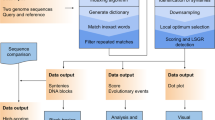
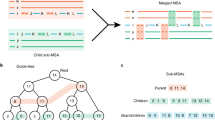
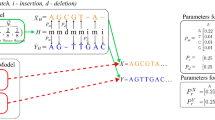
Abstract
Even though the recent advances in ‘complete genomics’ revealed the previously inaccessible genomic regions, analysis of variations in centromeres and other extra-long tandem repeats (ETRs) faces an algorithmic challenge since there are currently no tools for accurate sequence comparison of ETRs. Counterintuitively, the classical alignment approaches, such as the Smith–Waterman algorithm, fail to construct biologically adequate alignments of ETRs. We present UniAligner—the parameter-free sequence alignment algorithm with sequence-dependent alignment scoring that automatically changes for any pair of compared sequences. UniAligner prioritizes matches of rare substrings that are more likely to be relevant to the evolutionary relationship between two sequences. We apply UniAligner to estimate the mutation rates in human centromeres, and quantify the extremely high rate of large duplications and deletions in centromeres. This high rate suggests that centromeres may represent some of the most rapidly evolving regions of the human genome with respect to their structural organization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
We used the T2T assembly v2.0 of the CHM13 genome (available from https://s3-us-west-2.amazonaws.com/human-pangenomics/T2T/CHM13/assemblies/analysis_set/chm13v2.0.fa.gz); heavy immunoglobulin locus of human (IGHDH; NC_000014.9: 105,865,737–105,964,717) and of orangutan (NC_036917.1: 87,805,787–87,899,312); HG002 v0.7 assembly (publicly available at https://github.com/marbl/hg002). The test launch command on a small test dataset is available in the ‘Makefile’. Alignment of cenX1 and cenX2 generated by UniAligner is located at Zenodo69.
Code availability
The codebase of UniAligner is available at https://github.com/seryrzu/tandem_aligner.
The source code of version 0.1 that was used in this study is available at Zenodo69.
References
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Wang, T. et al. The Human Pangenome Project: a global resource to map genomic diversity. Nature 604, 437–446 (2022).
Liao, W.-W. et al. A draft human pangenome reference. Nature 617, 312–324 (2023).
Vollger, M. R. et al. Segmental duplications and their variation in a complete human genome. Science 376, eabj6965 (2022).
Hoyt, S. J. et al. From telomere to telomere: the transcriptional and epigenetic state of human repeat elements. Science 376, eabk3112 (2022).
Bakhtiari, M. et al. Variable number tandem repeats mediate the expression of proximal genes. Nat. Commun. 12, 2075 (2021).
Park, J., Bakhtiari, M., Popp, B., Wiesener, M. & Bafna, V. Detecting tandem repeat variants in coding regions using code-adVNTR. iScience 25, 104785 (2022).
Dvorkina, T., Kunyavskaya, O., Bzikadze, A. V., Alexandrov, I. & Pevzner, P. A. CentromereArchitect: inference and analysis of the architecture of centromeres. Bioinformatics 37, i196–i204 (2021).
Altemose, N. et al. Complete genomic and epigenetic maps of human centromeres. Science 376, eabl4178 (2022).
Kunyavskaya, O., Dvorkina, T., Bzikadze, A. V., Alexandrov, I. & Pevzner, P. A. Automated annotation of human centromeres with HORmon. Genome Res. 32, 1137–1151 (2022).
Schueler, M. G., Higgins, A. W., Rudd, M. K., Gustashaw, K. & Willard, H. F. Genomic and genetic definition of a functional human centromere. Science 294, 109–115 (2001).
Alkan, C. et al. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput. Biol. 3, 1807–1818 (2007).
Enukashvily, N. I., Donev, R., Waisertreiger, I. S.-R. & Podgornaya, O. I. Human chromosome 1 satellite 3 DNA is decondensed, demethylated and transcribed in senescent cells and in A431 epithelial carcinoma cells. Cytogenet. Genome Res. 118, 42–54 (2007).
Shepelev, V. A., Alexandrov, A. A., Yurov, Y. B. & Alexandrov, I. A. The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLoS Genet. 5, e1000641 (2009).
Nagaoka, S. I., Hassold, T. J. & Hunt, P. A. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504 (2012).
Melters, D. P. et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14, R10 (2013).
Giunta, S. & Funabiki, H. Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc. Natl Acad. Sci. USA 114, 1928–1933 (2017).
Black, E. M. & Giunta, S. Repetitive fragile sites: centromere satellite DNA as a source of genome instability in human diseases. Genes 9, 615 (2018).
Smurova, K. & De Wulf, P. Centromere and pericentromere transcription: roles and regulation … in sickness and in health. Front. Genet. https://doi.org/10.3389/fgene.2018.00674 (2018).
Miga, K. H. Centromeric satellite DNAs: hidden sequence variation in the human population. Genes 10, 352 (2019).
Miga, K. H. & Alexandrov, I. A. Variation and evolution of human centromeres: a field guide and perspective. Annu. Rev. Genet. 55, 583–602 (2021).
Sirupurapu, V., Safonova, Y. & Pevzner, P. A. Gene prediction in the immunoglobulin loci. Genome Res. 32, 1152–1169 (2022).
Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics 37, 4572–4574 (2021).
Ekim, B., Sahlin, K., Medvedev, P., Berger, B. & Chikhi, R. Efficient mapping of accurate long reads in minimizer space with mapquik. Genome Res. https://doi.org/10.1101/gr.277679.123 (2023).
Xu, C. A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput. Struct. Biotechnol. J. 16, 15–24 (2018).
Van der Auwera, G. A. & O’Connor, B. D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra (O’Reilly Media, 2020).
Bakhtiari, M., Shleizer-Burko, S., Gymrek, M., Bansal, V. & Bafna, V. Targeted genotyping of variable number tandem repeats with adVNTR. Genome Res. 28, 1709–1719 (2018).
Mousavi, N., Shleizer-Burko, S., Yanicky, R. & Gymrek, M. Profiling the genome-wide landscape of tandem repeat expansions. Nucleic Acids Res. 47, e90 (2019).
Chaisson, M. J. P. et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 10, 1784 (2019).
Bzikadze, A. V. & Pevzner, P. A. Automated assembly of centromeres from ultra-long error-prone reads. Nat. Biotechnol. 38, 1309–1316 (2020).
Nurk, S. et al. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res. 30, 1291–1305 (2020).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Bankevich, A., Bzikadze, A. V., Kolmogorov, M., Antipov, D. & Pevzner, P. A. Multiplex de Bruijn graphs enable genome assembly from long, high-fidelity reads. Nat. Biotechnol. 40, 1075–1081 (2022).
Rautiainen, M. et al. Telomere-to-telomere assembly of diploid chromosomes with Verkko. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01662-6 (2023).
Ekim, B., Berger, B. & Chikhi, R. Minimizer-space de Bruijn graphs: whole-genome assembly of long reads in minutes on a personal computer. Cell Syst. 12, 958–968 (2021).
Bickhart, D. M. et al. Generating lineage-resolved, complete metagenome-assembled genomes from complex microbial communities. Nat. Biotechnol. 40, 711–719 (2022).
Mikheenko, A., Bzikadze, A. V., Gurevich, A., Miga, K. H. & Pevzner, P. A. TandemTools: mapping long reads and assessing/improving assembly quality in extra-long tandem repeats. Bioinformatics 36, i75–i83 (2020).
Smith, T. F. & Waterman, M. S. Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197 (1981).
Miga, K. H. et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 585, 79–84 (2020).
Rudd, M. K., Wray, G. A. & Willard, H. F. The evolutionary dynamics of α-satellite. Genome Res. 16, 88–96 (2006).
Pertile, M. D., Graham, A. N., Choo, K. H. A. & Kalitsis, P. Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res. 19, 2202–2213 (2009).
Chaisson, M. J. & Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13, 238 (2012).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Manber, U. & Myers, G. Suffix arrays: a new method for on-line string searches. SIAM J. Comput. 22, 935–948 (1993).
Smith, G. P. Evolution of repeated DNA sequences by unequal crossover. Science 191, 528–535 (1976).
Gibbs, A. J. & McIntyre, G. A. The diagram, a method for comparing sequences. Its use with amino acid and nucleotide sequences. Eur. J. Biochem. 16, 1–11 (1970).
Vollger, M. R., Kerpedjiev, P., Phillippy, A. M. & Eichler, E. E. StainedGlass: interactive visualization of massive tandem repeat structures with identity heatmaps. Bioinformatics 38, 2049–2051 (2022).
Watson, C. T. & Breden, F. The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes Immun. 13, 363–373 (2012).
Rodriguez, O. L. et al. A novel framework for characterizing genomic haplotype diversity in the human immunoglobulin heavy chain locus. Front. Immunol. 11, 2136 (2020).
Rhie, A. et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 (2021).
Koonin, E. V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1, 127–136 (2003).
Safonova, Y. & Pevzner, P. A. V(DD)J recombination is an important and evolutionarily conserved mechanism for generating antibodies with unusually long CDR3s. Genome Res. 30, 1547–1558 (2020).
Šošić, M. & Šikić, M. Edlib: a C/C++ library for fast, exact sequence alignment using edit distance. Bioinformatics 33, 1394–1395 (2017).
Eppstein, D., Galil, Z., Giancarlo, R. & Italiano, G. F. Sparse dynamic programming I: linear cost functions. J. ACM 39, 519–545 (1992).
Arratia, R. & Waterman, M. S. A phase transition for the score in matching random sequences allowing deletions. Ann. Appl. Probab. 4, 200–225 (1994).
Waterman, M. S. & Vingron, M. Sequence comparison significance and Poisson approximation. Stat. Sci. 9, 367–381 (1994).
Altemose, N. et al. Complete genomic and epigenetic maps of human centromeres. Science 376, eabl4178 (2022).
Manber, U. & Myers, G. Suffix arrays: a new method for on-line string searches. SIAM J. Comput. 22, 935–948 (1993).
Kasai, T., Lee, G., Arimura, H., Arikawa, S. & Park, K. Linear-time longest-common-prefix computation in suffix arrays and its applications. In Combinatorial Pattern Matching (ed. Landau, G. M.) 181–192 (Springer, 2001).
Larsson, N. J. & Sadakane, K. Faster Suffix Sorting (Dept. of Computer Science, Lund Univ., 1999).
Burkhardt, S. & Kärkkäinen, J. Fast lightweight suffix array construction and checking. In Proc. 14th Annual Symposium on Combinatorial Pattern Matching (eds. Baeza-Yates, R. et al.) 55–69 (Springer, 2003).
Kärkkäinen, J. & Sanders, P. Simple linear work suffix array construction. In Lecture Notes in Computer Science (eds. Baeten, J. C. M. et al.) 943–955 (Springer, 2003).
Kim, D. K., Sim, J. S., Park, H. & Park, K. Linear-time construction of suffix arrays. In Proc 14th Annual Symposium on Combinatorial Pattern Matching (eds. Baeza-Yates, R. et al.) 186–199 (Springer, 2003).
Ko, P. & Aluru, S. Space efficient linear time construction of suffix arrays. J. Discrete Algorithms 3, 143–156 (2005).
Pearson, W. R. & Lipman, D. J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA 85, 2444–2448 (1988).
Logan, B. F. & Shepp, L. A. A variational problem for random Young tableaux. Adv. Math. 26, 206–222 (1977).
Vershik, A. M. & Kerov, S. V. Asymptotics of the Plancherel measure of the symmetric group and the limiting form of Young tableaux. Dokl. Akad. Nauk SSSR 233, 1024–1027 (1977).
Logsdon, G. A. et al. The structure, function and evolution of a complete human chromosome 8. Nature 593, 101–107 (2021).
Bzikadze, A. V. & Pevzner, P. A. UniAligner: a new parameter-free framework for fast sequence alignment. Zenodo https://doi.org/10.5281/zenodo.7563836 (2023).
Acknowledgements
We are indebted to K. Miga and M. Cechova for providing early pedigree centromere assemblies. We are grateful to I. Alexadrov, A. Bankevich, A.V. Bzikadze, R. Chikhi, T. Dvorkina, E. Eichler, G. Logsdon, O. Kunyavskaya and C. Wu for helpful discussions and suggestions. A.V.B. and P.A.P. were supported by the National Science Foundation EAGER award 2032783.
Author information
Authors and Affiliations
Contributions
A.V.B. conducted the experiments and wrote the code for UniAligner. P.A.P. supervised the research. All authors worked on the development of the UniAligner algorithm and wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Jue Ruan, Chengzhi Liang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Lei Tang and Lin Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Notes 1–14.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bzikadze, A.V., Pevzner, P.A. UniAligner: a parameter-free framework for fast sequence alignment. Nat Methods 20, 1346–1354 (2023). https://doi.org/10.1038/s41592-023-01970-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-023-01970-4