Abstract
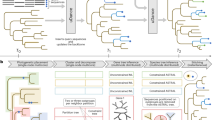
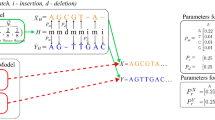
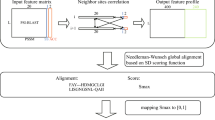
Multiple sequence alignments (MSAs) are used for structural1,2 and evolutionary predictions1,2, but the complexity of aligning large datasets requires the use of approximate solutions3, including the progressive algorithm4. Progressive MSA methods start by aligning the most similar sequences and subsequently incorporate the remaining sequences, from leaf to root, based on a guide tree. Their accuracy declines substantially as the number of sequences is scaled up5. We introduce a regressive algorithm that enables MSA of up to 1.4 million sequences on a standard workstation and substantially improves accuracy on datasets larger than 10,000 sequences. Our regressive algorithm works the other way around from the progressive algorithm and begins by aligning the most dissimilar sequences. It uses an efficient divide-and-conquer strategy to run third-party alignment methods in linear time, regardless of their original complexity. Our approach will enable analyses of extremely large genomic datasets such as the recently announced Earth BioGenome Project, which comprises 1.5 million eukaryotic genomes6.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data, analyses and results are available from Zenodo (https://doi.org/10.5281/zenodo.3271452).
Code availability
The regressive alignment algorithm has been implemented in T-Coffee and is available at the T-Coffee website (http://www.tcoffee.org) and on GitHub (https://github.com/cbcrg/tcoffee). A GitHub repository containing the Nextflow workflow25 and Jupyter notebooks26 to replicate the analysis are available at https://github.com/cbcrg/dpa-analysis (release v.1.2).
References
Uguzzoni, G. et al. Large-scale identification of coevolution signals across homo-oligomeric protein interfaces by direct coupling analysis. Proc. Natl Acad. Sci. USA 114, E2662–E2671 (2017).
Mirarab, S., Bayzid, M. S., Boussau, B. & Warnow, T. Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 346, 1250463 (2014).
Wang, L. & Jiang, T. On the complexity of multiple sequence alignment. J. Comput. Biol. 1, 337–348 (1994).
Hogeweg, P. & Hesper, B. The alignment of sets of sequences and the construction of phylogenetic trees. An integrated method. J. Mol. Evol. 20, 175–186 (1984).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Lewin, H. A. et al. Earth BioGenome Project: sequencing life for the future of life. Proc. Natl Acad. Sci. USA 115, 4325–4333 (2018).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Chatzou, M. et al. Multiple sequence alignment modeling: methods and applications. Brief. Bioinform. 17, 1009–1023 (2015).
Breen, M. S., Kemena, C., Vlasov, P. K., Notredame, C. & Kondrashov, F. A. Epistasis as the primary factor in molecular evolution. Nature 490, 535–538 (2012).
Liu, K., Raghavan, S., Nelesen, S., Linder, C. R. & Warnow, T. Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science 324, 1561–1564 (2009).
Mirarab, S. et al. PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J. Comput. Biol. 22, 377–386 (2015).
Collins, K. & Warnow, T. PASTA for proteins. Bioinformatics 34, 3939–3941 (2018).
Nguyen, N.-P. D., Mirarab, S., Kumar, K. & Warnow, T. Ultra-large alignments using phylogeny-aware profiles. Genome Biol. 16, 124 (2015).
Yamada, K. D., Tomii, K. & Katoh, K. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics 32, 3246–3251 (2016).
Minh, B. Q., Klaere, S. & von Haeseler, A. Phylogenetic diversity within seconds. Syst. Biol. 55, 769–773 (2006).
Stebbings, L. A. & Mizuguchi, K. HOMSTRAD: recent developments of the homologous protein structure alignment database. Nucleic Acids Res. 32, D203–D207 (2004).
Blackshields, G., Sievers, F., Shi, W., Wilm, A. & Higgins, D. G. Sequence embedding for fast construction of guide trees for multiple sequence alignment. Algorithms Mol. Biol. 5, 21 (2010).
Katoh, K. & Toh, H. PartTree: an algorithm to build an approximate tree from a large number of unaligned sequences. Bioinformatics 23, 372–374 (2007).
Katoh, K., Kuma, K.-I., Toh, H. & Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005).
Greenacre, M. J. Biplots in Practice (Fundacion BBVA, 2010).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2013).
Herlihy, M. & Shavit, N. The Art of Multiprocessor Programming 1st edn (Morgan Kaufmann, 2012).
Kahn, S. D. On the future of genomic data. Science 331, 728–729 (2011).
Thompson, J. D., Plewniak, F. & Poch, O. BAliBASE: a benchmark alignment database for the evaluation of multiple alignment programs. Bioinformatics 15, 87–88 (1999).
Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Perkel, J. M. Why Jupyter is data scientists’ computational notebook of choice. Nature 563, 145–146 (2018).
Acknowledgements
We thank G. Riddihough for revisions and comments on the manuscript and O. Gascuel for suggestions. This project was supported by the Centre for Genomic Regulation, the Spanish Plan Nacional, the Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa’ (E.G., P.T., C.M., I.E., L.M., A.B., F.K., E.F. and C.N.) and an ERC Consolidator Grant from the European Commission, grant agreement no. 771209 ChrFL (F.K.).
Author information
Authors and Affiliations
Contributions
C.N. designed and implemented the algorithm. E.F., E.G., L.M., A.B. and P.D.T designed the validation procedure and carried out the validation. I.E. performed statistical and CCA analyses. E.F., C.N., E.G., C.M., L.M., A.B., P.D.T., I.E., F.K. and H.L. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Effect of size of N on HomFam average TC score and relative CPU time.
Summary of Total Column score and CPU usage values collected over HomFam datasets of various sizes of N using either (A) ClustalO with mBed trees or (B) Mafft-FFTNS1 with PartTree trees. Four datasets were excluded from the analysis because of incomplete CPU time reports across the runs (mmp, kunitz, hormone_rec and peroxidase). Each combination of alignment method and tree method, n=90 independent MSA samples.
Supplementary Figure 2 Relative performances of alternative MSA algorithm combinations.
(A) The relative accuracy is defined as the difference between the TC score measured on the projection of embedded sequences and the TC score measured on the direct alignment of these same sequences with the considered method. The three alignment protocols all use a PartTree guide-trees combined with the following aligners Fftns1 in non-regressive mode (red), Fftns1 in regressive mode (green) and Gins1 in regressive mode (blue). The envelope is the standard deviation measured on the averaged values. (B) similar comparison between the regressive deployment of Sparsecore using a mBed guide tree (blue) and the default, non-regressive deployment of this same aligner (red). (C) similar comparison on UPP using a mBed guide-tree for the regressive deployment (blue) and UPP default mode for the non-regressive (red). (D) similar display for Fftns1 using a mBed guide-tree for the regressive (blue) and non-regressive deployment (red). (E) similar analysis using ClustalO as an aligner and PartTree guide-trees in regressive (blue) and non-regressive modes (red). Each combination of alignment method and tree method, n=94 independent MSA samples.
Supplementary information
Supplementary Materials
Supplementary Figs. 1 and 2, Notes 1 and 2 and Tables 1–7.
Rights and permissions
About this article
Cite this article
Garriga, E., Di Tommaso, P., Magis, C. et al. Large multiple sequence alignments with a root-to-leaf regressive method. Nat Biotechnol 37, 1466–1470 (2019). https://doi.org/10.1038/s41587-019-0333-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0333-6
This article is cited by
-
Phylogenetic and functional characterization of water bears (Tardigrada) tubulins
Scientific Reports (2023)
-
Reference flow: reducing reference bias using multiple population genomes
Genome Biology (2021)