Abstract
For patients with non-small-cell lung cancer (NSCLC) tumors without currently targetable molecular alterations, standard-of-care treatment is immunotherapy with anti-PD-(L)1 checkpoint inhibitors, alone or with platinum-doublet therapy. However, not all patients derive durable benefit and resistance to immune checkpoint blockade is common. Understanding mechanisms of resistance—which can include defects in DNA damage response and repair pathways, alterations or functional mutations in STK11/LKB1, alterations in antigen-presentation pathways, and immunosuppressive cellular subsets within the tumor microenvironment—and developing effective therapies to overcome them, remains an unmet need. Here the phase 2 umbrella HUDSON study evaluated rational combination regimens for advanced NSCLC following failure of anti-PD-(L)1-containing immunotherapy and platinum-doublet therapy. A total of 268 patients received durvalumab (anti-PD-L1 monoclonal antibody)–ceralasertib (ATR kinase inhibitor), durvalumab–olaparib (PARP inhibitor), durvalumab–danvatirsen (STAT3 antisense oligonucleotide) or durvalumab–oleclumab (anti-CD73 monoclonal antibody). Greatest clinical benefit was observed with durvalumab–ceralasertib; objective response rate (primary outcome) was 13.9% (11/79) versus 2.6% (5/189) with other regimens, pooled, median progression-free survival (secondary outcome) was 5.8 (80% confidence interval 4.6–7.4) versus 2.7 (1.8–2.8) months, and median overall survival (secondary outcome) was 17.4 (14.1–20.3) versus 9.4 (7.5–10.6) months. Benefit with durvalumab–ceralasertib was consistent across known immunotherapy-refractory subgroups. In ATM-altered patients hypothesized to harbor vulnerability to ATR inhibition, objective response rate was 26.1% (6/23) and median progression-free survival/median overall survival were 8.4/22.8 months. Durvalumab–ceralasertib safety/tolerability profile was manageable. Biomarker analyses suggested that anti-PD-L1/ATR inhibition induced immune changes that reinvigorated antitumor immunity. Durvalumab–ceralasertib is under further investigation in immunotherapy-refractory NSCLC.
ClinicalTrials.gov identifier: NCT03334617
Similar content being viewed by others
Main
Patients with advanced non-small-cell lung cancer (NSCLC) receive initial therapy based on molecular classification of their disease1. For NSCLC tumors without currently targetable molecular alterations2 (>50% of adenocarcinomas3 and >95% of squamous cell carcinomas4), standard-of-care treatment is immunotherapy with anti-programmed death (ligand)-1 (PD-(L)1) checkpoint inhibitors, alone or with platinum-doublet therapy1, which offers long-term disease control and overall survival (OS) benefit for some patients5,6,7,8,9,10,11,12. Tumor-intrinsic factors and the tumor microenvironment (TME) influence response to immune checkpoint blockade (ICB)13,14,15,16, including genomic features associated with differential response to ICB. Although high PD-L1 expression levels7,17 and tumor mutational burden (TMB)18 have emerged as important biomarkers, not all patients derive durable benefit, and resistance to ICB is common. Clinical resistance to ICB is complex and can present at various time points during treatment; primary resistance is defined as a best response of disease progression or stable disease lasting <6 months (24 weeks) and acquired resistance as progression following a response or stable disease lasting ≥6 months13,19,20,21. Biological mechanisms that render patients with NSCLC recalcitrant to ICB include alterations or functional mutations resulting in the inactivation of the tumor suppressor gene STK11/LKB1 and a consequent reduction in tumor-infiltrating lymphocytes22,23,24,25, inactivating mutations of KEAP1 (refs. 26,27), low expression of PD-L1, and alterations in antigen-presentation pathways. Furthermore, immunosuppressive cellular subsets within the TME such as myeloid-derived suppressor cells (MDSCs), T regulatory cells and others influence responses to ICB16. Understanding these resistance mechanisms, and developing effective therapies to overcome them, remains a critical unmet need as there are currently no immunotherapy-based regimens approved for patients with NSCLC who have progressed after initial ICB.
A major challenge in this setting is our poor understanding of the key immunotherapy-resistance mechanisms in primary and acquired resistance. While genotype/biomarker-directed targeted therapy is relatively well understood and can offer durable efficacy in NSCLC28,29,30, less is understood about biomarker subgroups in the immunotherapy setting16 that may be associated with reduced efficacy. The ongoing, open-label, multicenter, nonrandomized, modular, phase 2 umbrella HUDSON study (NCT03334617) was designed to build a comprehensive understanding of key features in patients and their tumors associated with disease progression and to differentiate tumor characteristics and treatment activity in primary and acquired resistance phenotypes. Rational combination therapies were selected for investigation in HUDSON based on targeting proposed mechanisms of immunosuppression with the aim of reinvigorating immune-mediated antitumor activity.
Mismatch repair deficiency is associated with greater benefit from immunotherapies31,32,33,34; HUDSON therefore evaluated inhibitors of DNA damage response (DDR) and repair pathways in combination with the anti-PD-L1 monoclonal antibody durvalumab. DDR pathway defects generate immunogenic neoantigens that are recognized and targeted by T cells32,35,36. Inhibition of poly-(ADP-ribose) polymerase (PARP)37,38,39 and ataxia telangiectasia and Rad3-related (ATR) protein kinase40,41 represent two key approaches for targeting DDR defects, and HUDSON evaluated both PARP and ATR inhibitors in combination with durvalumab. Another approach may be to target immunosuppression in the TME and resistance lymph nodes. For example, signal transducer and activator of transcription 3 (STAT3) signaling results in production of tumor-promoting cytokines (by myeloid cells in the TME) and inhibition of antitumor cytokines, shifting the TME toward an immunosuppressive state42. Targeting STAT3 may help reverse immunosuppression by myeloid cells and directly attack tumor cells (TCs)43. Similarly, targeting anti-5′-nucleotidase cluster of differentiation 73 (CD73) may help reverse the immunosuppressive TME by increasing the activity of CD8-positive effector cells, activating macrophages and reducing both MDSCs and regulatory T lymphocytes44,45.
In this Article, we report the first clinical efficacy, safety, and translational data from four modules of HUDSON in patients who have received a platinum-doublet regimen and progressed on anti-PD-(L)1-based therapy. Treatments received in these modules were durvalumab plus ceralasertib (ATR kinase inhibitor), olaparib (PARP inhibitor), danvatirsen (STAT3-targeting antisense oligonucleotide) or oleclumab (anti-CD73 monoclonal antibody). Patients underwent tumor molecular profiling at screening and were assigned to either a biomarker-matched cohort (durvalumab–ceralasertib: ATM altered; durvalumab–olaparib: homologous recombination repair (HRR) altered, STK11/LKB1 altered; durvalumab–oleclumab: high CD73 expression) or a biomarker-nonmatched group. Patients in the latter group were further stratified into primary or acquired resistance cohorts (see ‘Study design’ in Methods). Multi-omic longitudinal peripheral biomarker profiling was used for hypothesis-generating exploratory analyses of possible underlying mechanisms of action driving treatment outcomes.
Results
Patients
Between 26 January 2018 and 26 April 2022, 941 patients were screened for enrollment in HUDSON, including 324 for the four modules reported herein (Fig. 1); of these, 268 patients received treatment, 88 within biomarker-matched cohorts and 77 and 103 within primary and acquired resistance biomarker-nonmatched cohorts, respectively. Overall, 79 patients received durvalumab–ceralasertib and 189 received the other regimens, including 87, 45 and 57 who received durvalumab–olaparib, durvalumab–danvatirsen and durvalumab–oleclumab, respectively. Patient demographics and disease characteristics were similar between treatment modules (Table 1).
Based on tumor molecular profiling, patients were assigned to either biomarker-matched (Group A) or non-matched cohorts (Group B), which included patients with primary and acquired resistance determined by their initial response to prior immunotherapy-containing regimens. Patients were then treated with one of four durvalumab-based combination regimens (Modules 1–4).
Notable clinical efficacy with ceralasertib–durvalumab
Efficacy was evaluated with the four rational combinations to determine whether targeting specific pathways potentially associated with resistance could improve outcomes. Objective response rate (ORR), the primary endpoint, was 13.9% (n = 11/79) with durvalumab–ceralasertib, whereas the pooled ORR across the other treatment modules was 2.6% (n = 5/189) (Extended Data Table 1). All objective responses were confirmed partial responses (see Supplementary Table 1 for details of prior treatment in responding patients). Disease control rates at 12 and 24 weeks, respectively, were 50.6% and 35.4% with durvalumab–ceralasertib and 32.3% and 15.9% with the pooled other regimens (Extended Data Table 1).
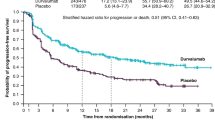
Longer progression-free survival (PFS) and OS were seen with durvalumab–ceralasertib compared with the pooled other regimens, with a median PFS of 5.8 (80% confidence interval (CI) 4.6–7.4) and 2.7 (1.8–2.8) months, respectively, and a median OS of 17.4 (14.1–20.3) and 9.4 (7.5–10.6) months, respectively (Fig. 2a, b and Extended Data Table 1), after median follow-up times in censored patients of 8.3 and 28.2 months.
Durvalumab–ceralasertib in ATM biomarker-matched patients
In patients with genetic or immunosuppressive biomarkers of relevance, the mechanisms of action of the partner drugs were investigated to determine the efficacy and specific sensitivity of durvalumab-based combinations in these settings. Patient demographics and baseline disease characteristics in the biomarker-matched and biomarker-nonmatched groups are summarized in Supplementary Table 2. Responses and outcomes by treatment module and in individual biomarker-matched and primary and acquired biomarker-nonmatched cohorts are summarized in Extended Data Table 2 for durvalumab–ceralasertib and in Supplementary Tables 3–5 for durvalumab–olaparib, durvalumab–danvatirsen and durvalumab–oleclumab. Kaplan–Meier analyses of PFS and OS by cohort are shown in Extended Data Fig. 1.
ATM alterations confer ATR dependency in tumors. We observed an ORR of 26.1% (n = 6/23) with durvalumab–ceralasertib in the ATM-altered biomarker-matched cohort, higher than the ORRs of 13.0% (n = 3/23) and 6.1% (n = 2/33) in the primary and acquired resistance biomarker-nonmatched cohorts, respectively (Extended Data Table 2), suggesting specific benefit of ATR inhibition in patients with ATM alterations. Additionally, PFS (median 8.4 versus 4.6 versus 4.6 months) and OS (median 22.8 versus 12.0 versus 19.1 months) appeared longer in the ATM-altered biomarker-matched cohort versus the primary and acquired resistance cohorts, respectively (Extended Data Fig. 1a,b). Notably, while OS appeared longer in the acquired versus primary resistance cohorts, PFS was similar.
In contrast, PARP inhibition has been successful in targeting tumors with DDR defects in other solid tumors, and preclinical data suggest that STK11/LKB1 alterations may be associated with enhanced PARP inhibitor sensitivity25,46. However, ORR with durvalumab–olaparib was 4.6%, including 9.5% and 4.8% in the HRRm and STK11/LKB1 biomarker-matched cohorts, suggesting limited vulnerability from targeting these mutations, and 0% and 4.3% in the primary and acquired resistance cohorts (Supplementary Table 3). Similarly, no objective responses were seen with durvalumab–danvatirsen (Supplementary Table 4), indicating that reprogramming macrophages with STAT3 inhibition using an antisense oligonucleotide does not reverse myeloid-mediated resistance, even in the context of STK11 alterations, which have been associated with suppression of mononuclear and polymorphonuclear MDSCs22,26. One partial response (1.8%) was seen with durvalumab–oleclumab (Supplementary Table 5), suggesting that reducing adenosine levels through CD73 inhibition is insufficient to reverse CD73-dependent immunosuppression. PFS and OS with durvalumab–olaparib (Extended Data Fig. 1c,d) and durvalumab–oleclumab (Extended Data Fig. 1g,h) appeared similar or shorter in the biomarker-matched versus biomarker-nonmatched cohorts, with PFS and OS with each regimen also appearing shorter in the biomarker-nonmatched primary versus acquired resistance cohorts (Extended Data Fig. 1c–h).
Treatment exposure and safety
Mean durvalumab treatment duration was longer with durvalumab–ceralasertib (8.7 months) than the other regimens (5.1 months), and this was reflected across cohorts (Extended Data Table 3). With durvalumab–ceralasertib and the other regimens, similar overall incidences of treatment-emergent adverse events (TEAEs; 93.7% and 89.9%), grade ≥3 TEAEs (44.3% and 51.3%), treatment-related TEAEs (TRAEs; 75.9% and 71.4%), grade ≥3 TRAEs (20.3% and 30.2%) and serious AEs (SAEs; 36.7% and 34.4%) were reported (Extended Data Table 4). Incidences of treatment-related SAEs (12.7% and 9.0%) and treatment discontinuation due to TRAEs (7.6% and 10.1%) were low and similar with durvalumab–ceralasertib and the other regimens, respectively. Two patients (2.5%) receiving durvalumab–ceralasertib (pneumonia and myocardial infarction, each n = 1) and five patients (2.6%) receiving the other regimens (cor pulmonale, dyspnea, sepsis, pneumonia aspiration and renal artery thrombosis, each n = 1), died due to a TEAE; none was considered related to treatment. Safety profiles by regimen, overall and by cohort, are summarized in Supplementary Tables 6–9.
Among Medical Dictionary for Regulatory Activities (MedDRA) System Organ Class (SOC) categories, the incidences of gastrointestinal disorders (58.2% and 34.9%), metabolism and nutrition disorders (25.3% and 10.1%) and nervous system disorders (19.0% and 4.8%) showed >10 percentage point differences between durvalumab–ceralasertib and the other regimens, pooled; of the individual TRAEs occurring in ≥10% of patients treated with durvalumab–ceralasertib or other regimens, nausea (50.6% and 22.2%), vomiting (27.8% and 11.1%) and decreased appetite (21.5% and 7.4%) were the only TRAEs for which there was a >10 percentage point difference between groups (Extended Data Table 4). Incidences of TRAEs by MedDRA SOC and of individual TRAEs, and common grade ≥3 TRAEs by individual regimen, are summarized in Extended Data Tables 5 and 6, respectively. With durvalumab–ceralasertib, the only grade ≥3 TRAE with an incidence of ≥5% was thrombocytopenia (5.1%), while with durvalumab–olaparib anemia was reported in 13.8% of patients, and with durvalumab–danvatirsen increased alanine aminotransferase was reported in 11.1% and thrombocytopenia in 8.9% of patients (Extended Data Table 6).
Distinct features delineate patients with ICB-recalcitrant NSCLC
In an exploratory post-hoc analysis to examine clinical and molecular biomarker characteristics associated with recalcitrance to ICB, we measured TME inflammation status using a validated 18-gene tumor inflammation signature (TIS)47,48 in subgroups with known association with poor immunotherapy response among patients with evaluable prescreening tumor biopsy suitable for bulk-RNA sequencing who received durvalumab plus olaparib, danvatirsen or oleclumab (Supplementary Table 10). This composite signature, which reflects the state, function and phenotype of the TME and is a widely used predictive biomarker for benefit with ICB therapy47,49,50, was used to characterize TME immune and inflammation status in patient subgroups in which clinical data for durvalumab–ceralasertib and durvalumab plus olaparib, danvatirsen or oleclumab were evaluated.
TIS scores (with pooled OS data among all patients who received durvalumab plus olaparib, danvatirsen or oleclumab, for context) are shown in Fig. 3. OS was numerically longer and TIS score higher among patients with acquired versus primary resistance, with multiple markers of inflammation including lymphocyte-activation gene 3 (LAG3), T cell immunoreceptor with immunoglobulin and immunoreceptor tyrosine-based inhibitory motif (ITIM) domains (TIGIT) and nonclassical human leukocyte antigen (HLA) molecule HLA-E more highly expressed in those with acquired resistance to prior immunotherapy (Fig. 3a and Extended Data Fig. 2a). Analysis according to PD-(L)1 TC expression status (<1%, negative, versus ≥1%, positive) showed similar outcomes in both groups and a similar TIS score (Fig. 3b) but with elevated expression of some inflammation markers in some PD-(L)1-positive tumors (Extended Data Fig. 2b). OS appeared longer and TIS score was higher (P = 0.0517) in patients with adenocarcinoma versus squamous cell histology, with most markers of inflammation more highly expressed in adenocarcinomas (Fig. 3c). Similarly, the presence of bone and/or liver metastases was associated with shorter OS; however, there was no difference in TIS score between those with or without metastases, albeit sites of biopsy differed and may have impacted the findings (Fig. 3d). Outcomes and TIS score were broadly similar in subgroups with high or low TMB, and altered or wild-type STK11 (Fig. 3e,f). Heat maps showing expression of the 18 genes in the TIS in the above subgroups are shown in Extended Data Fig. 2.
a–f, Kaplan–Meier distributions of OS (left) and TIS ssGSEA enrichment score (ES) distributions (violin plots, right) comparing patients with primary or acquired resistance (a), PD-L1-negative or PD-L1-positive status (b), squamous cell carcinoma or adenocarcinoma histology (c), with or without bone/liver metastases (d), high or low TMB (e) and STK11-altered or STK11-wild-type tumors (f). Median OS, 80% CIs and numbers of patients at risk are shown in each Kaplan–Meier plot. Violin plots show data from individual patients (dots), color-coded by site of sample, the median value (text box) and the probability density distribution smoothed using a kernel density estimator (shaded areas). ssGSEA ES distributions were compared using two-sided Wilcoxon rank-sum tests, with no correction for multiplicity of testing; associated P values and the number of tumor samples in each group are reported for each comparison.
Durvalumab–ceralasertib broadly active in patient subgroups
Given the encouraging observations with durvalumab–ceralasertib versus other regimens, we evaluated outcomes in subgroups defined by molecular and tumor-related features to determine whether specific factors were associated with benefit from, or resistance to, durvalumab–ceralasertib. Demographics and baseline disease characteristics in patients treated with durvalumab–ceralasertib or the other regimens by primary or acquired resistance and by histology are summarized in Supplementary Tables 11 and 12, respectively. In patients with primary resistance, median PFS (6.0 versus 1.8 months) and OS (12.6 versus 6.3 months) were longer with durvalumab–ceralasertib compared with the other regimens (Fig. 4 and Extended Data Fig. 3a), with nonoverlapping 80% CIs, with similar findings in patients with acquired resistance (median PFS 5.8 versus 2.8 months; median OS 19.1 versus 12.6 months) (Fig. 4 and Extended Data Fig. 3b). PFS and OS were also consistently longer in patients with PD-L1-negative (median PFS 7.5 versus 2.1 months; median OS 20.3 versus 10.7 months) or PD-L1-positive (median PFS 4.6 versus 3.0 months; median OS 14.2 versus 9.7 months) tumors (Fig. 4 and Extended Data Fig. 3c,d), and in patients with squamous (median PFS 4.8 versus 2.6 months; median OS 15.9 versus 6.0 months) or nonsquamous (median PFS 6.0 versus 2.7 months; median OS 18.8 versus 10.8 months) histology (Fig. 4 and Extended Data Fig. 3e,f). Similar findings were seen across all recalcitrant subgroups (Fig. 4), including those with liver or bone metastases, low TMB, and variant STK11 (Extended Data Fig. 3g) and KRAS.
Kaplan–Meier analysis of PFS and OS with durvalumab–ceralasertib in subgroups defined by the presence or absence of the adverse prognostic factors described above (Extended Data Fig. 4) indicated that outcomes were similar in patients with primary or acquired resistance, PD-L1-negative or PD-L1-positive tumors, squamous or nonsquamous tumor histology, high or low TMB, and STK11 or KRAS mutant or wild-type tumors. Presence of liver or bone metastases was associated with somewhat shorter PFS and OS, but benefit compared with the other durvalumab-based regimens was nevertheless evident in this setting (Fig. 4). Overall, these findings indicate that tumors with features commonly associated with immunotherapy resistance are nevertheless sensitive to durvalumab–ceralasertib.
We also explored differential gene alterations between patients with a PFS of ≥6 or <6 months on durvalumab–ceralasertib in the biomarker-matched (ATM altered) and biomarker-nonmatched (ATM wild-type) cohorts (Supplementary Fig. 1). A difference in alteration frequency was observed for CDKN2A alterations, which were more common in patients with PFS <6 months (n = 4/10, 40.0%) than in patients with PFS ≥6 months (n = 0/11) in the biomarker-matched cohort. The small sample sizes in these analyses limit interpretation of the findings.
Ceralasertib immunomodulation enhances durvalumab effects
Pretreatment and on-treatment blood biomarkers were evaluated in exploratory hypothesis-generating analyses using multi-omic approaches to assess potential underlying mechanisms of action with durvalumab–ceralasertib treatment (Fig. 5). Longitudinal matched blood samples were collected at baseline, after 7 days of ceralasertib therapy and after durvalumab treatment (Fig. 5a) to evaluate gene expression and T cell receptor (TCR) profile dynamics in 48 and 62 patients, respectively (patient demographics and disease characteristics in Supplementary Table 13). Gene expression analysis revealed dynamic, reversible changes following 7 days of ceralasertib and before the first dose of durvalumab. In addition, changes in innate and adaptive immunity-relevant signatures51,52 were seen, including decreases in monocyte lineage and CD8 T-cell-associated and dysfunctional/exhausted T-cell-associated signatures, and increases in the hallmark tumor necrosis factor (TNF)-α-associated, interferon-γ response-associated and interferon-α response-associated signatures (Fig. 5b–g).
a, Schema depicting treatment schedule for modules 1 (durvalumab–olaparib), 2 (durvalumab–danvatirsen) and 3 (durvalumab–ceralasertib), showing durvalumab administration (blue arrows), combination agent dosing (continuous colored bars), and pretreatment/on-treatment blood sample collection time points (stars). BID, twice daily. b–g, Effects of ceralasertib and durvalumab treatment on longitudinal blood-derived transcriptomes are depicted in the box plots showing differentially expressed monocyte-associated signatures (b), CD8 T-cell-associated signatures (c), dysfunctional/exhausted CD8 T-cell-associated signatures (d), TNF-α-associated signatures (e), interferon-γ-associated signatures (f) and interferon-α-associated signatures (g). P values are for comparisons with respective cycle 0, day (D) 1 time points. P values for visit dates represent linear mixed model effects, illustrating changes in on-treatment samples. h–q, Effects of ceralasertib, olaparib and danvatirsen with durvalumab on longitudinal blood-derived T cell repertoire: box plots showing productive Simpson T cell clonality with durvalumab–ceralasertib (h); waterfall plots showing changes in clonality on treatment by patient with durvalumab–ceralasertib on cycle 0, day 1 versus cycle 1, day 1 (i) and cycle 1, day 1 versus cycle 1, day 22 (j); box plots showing distributions of T cell clonality with durvalumab–olaparib (k), and waterfall plots showing changes in clonality on treatment by patient with durvalumab–olaparib on cycle 1, day 1 versus cycle 1, day 15 (l); box plots showing distributions of T cell clonality with durvalumab–danvatirsen (m), and waterfall plots showing changes in clonality on treatment per patient with durvalumab–danvatirsen on cycle 1, day 1 versus cycle 1, day 15 (n); box plots of expanded T cell clones (o), newly detected expanded T cell clones (p) and Morisita index for durvalumab plus olaparib, danvatirsen or ceralasertib at indicated visit dates (q). P values for visit dates represent two-sided Wilcoxon paired signed-rank tests between time points or two-sided Wilcoxon signed-rank tests between study modules, with correction for multiplicity of testing (Benjamini–Hochberg procedure), illustrating changes in on-treatment samples. r, Proposed immunomodulatory mechanism of action effects of ceralasertib with durvalumab in the periphery and tumor. For all box plots (b–h, k, m and o–q), box centerlines show medians, box limits show upper and lower quartiles, and whiskers show range.
Longitudinal TCR sequencing demonstrated cyclical changes including, most notably, a reduction in clonality after 7 days of ceralasertib (P < 0.0001), with a return to baseline after durvalumab treatment (Fig. 5h,i). An increase in peripheral TCR clonality was observed in most patients (n = 48/58) after the addition of durvalumab (Fig. 5h,j); similar changes in T cell clonality were not observed in samples from patients receiving durvalumab–olaparib (Fig. 5k,l) or durvalumab–danvatirsen (Fig. 5m,n). An increase in peripheral T cell clonality in patients receiving durvalumab–ceralasertib was followed by an increase in expanded clones (P < 0.0001) (Fig. 5o) and newly detected expanded T cell clones (P < 0.0001) (Fig. 5p) after the addition of durvalumab. The net result of changes in the circulating T cell repertoire after ceralasertib followed by durvalumab treatment was an overall increase in clonality in most patients, but without a significant difference in the composition of the most abundant T cell clones, as measured by the Morisita index (Fig. 5q). Overall, in peripheral blood, signatures indicating decreases in exhausted T cells and, conversely, increased interferon pathway activation were observed in the on-ceralasertib treatment period. Furthermore, we observed enhanced expansion and maintenance of abundant T cell clones indicative of antitumor response. These results suggest a complementary mechanism of action whereby ceralasertib induces systemic immunomodulation that may be indicative of an enhanced antitumor immune response in combination with durvalumab (Fig. 5r).
Discussion
Critical challenges in the treatment of advanced NSCLC include identifying resistance mechanisms and finding new treatment options for use following failure of all standard-of-care therapies. For patients without targetable molecular alterations, this comprises treatment with platinum-doublet therapy and immunotherapy with an anti-PD-1/PD-L1 checkpoint inhibitor. Currently, there are few options offering clinical benefit for patients following failure of standard-of-care therapies31; therefore, one of the key aims of the HUDSON study is to identify regimens that can overcome inherent immune resistance in this setting. With its innovative, modular study design incorporating patient cohorts with specific biomarkers as well as biomarker-nonmatched cohorts with primary or acquired resistance, coupled with comprehensive characterization of key gene alterations via biological analysis of tumor material, HUDSON has enabled evaluation of multiple rational combinations and investigation of specific biomarkers associated with resistance.
Tumor-intrinsic factors and inflammation influence responses to ICB13,14,15,16. Higher TIS score is associated with anti-PD-(L)1 treatment benefit49 in the context of more substantial TME inflammation. We observed numerically higher TIS scores in subgroups with acquired resistance and adenocarcinoma histology compared with more ‘immune-cold’ subgroups with primary resistance and squamous cell histology. We found no differences in TIS score according to TMB or STK11 status; this contrasts with published findings49,53 and may be associated with differences in biology between the post-ICB samples in HUDSON and ICB-naive samples in prior publications. Of note, liver and bone metastases are generally associated with immune-cold features; however, the biopsies we analyzed were not limited to these metastatic sites, which may explain the lack of association with inflammation in this subgroup of patients.
This first report from HUDSON demonstrates a notable efficacy signal with durvalumab–ceralasertib, with substantially higher response and disease control rates and substantially longer PFS and OS than with the other regimens, pooled. Furthermore, the median PFS and OS with durvalumab–ceralasertib of 5.8 and 17.4 months, respectively, are notable in the context of reports from prospective clinical trials in previously treated NSCLC of docetaxel as second- or third-line therapy following platinum-based chemotherapy and immunotherapy (median PFS and OS of 4.0 and 10.5 months in the CONTACT-01 study54; median OS of 8.3 months with docetaxel or pemetrexed (81%/19%) in the ATALANTE-1 study55; median PFS and OS of 4.5 and 11.3 months in primarily nonsquamous NSCLC in the CodeBreaK 200 study56) or following platinum-based chemotherapy (median PFS and OS ranges of 2.8–4.2 and 8.1–9.9 months, respectively57,58,59,60,61), in which findings were better than or similar to the data for the other HUDSON regimens, pooled. These outcomes indicate that durvalumab–ceralasertib warrants further investigation in this NSCLC patient population with a high unmet need for new treatment options62.
Durvalumab–ceralasertib had greater activity in the ATM-altered biomarker-matched cohort, with higher response rates and longer outcomes compared with biomarker-nonmatched cohorts. This observation is consistent with the study’s predefined hypothesis that ATM-altered patients would derive greater benefit from ATR inhibition with ceralasertib, associated with the known dependence of ATM-altered tumors on ATR signaling63. It should be noted that, while data were mature for the majority of patients of the reported modules, follow-up was limited for a small number of more recently enrolled patients receiving durvalumab–ceralasertib, some of whom were not evaluable for response and/or had not reached the 12-week time point at data cutoff. Nevertheless, findings from the ATM biomarker-matched cohort reflect the promising antitumor activity, notably in patients with alterations in the DDR pathway, seen in a study of durvalumab–ceralasertib in patients with advanced/metastatic melanoma following anti-PD-1 failure64. By contrast, the hypothesis of greater benefit with durvalumab plus the PARP inhibitor olaparib in patients with HRR mutations or STK11 alterations did not appear to be borne out by our findings. Similarly, we did not see the hypothesized activity of durvalumab in combination with the STAT3 antisense oligonucleotide danvatirsen targeting the immunosuppressive TME or with the anti-CD73 monoclonal antibody oleclumab in patients with high CD73 expression.
Our subgroup analyses demonstrated broad activity with durvalumab–ceralasertib compared with the other regimens, including in patients with primary resistance, PD-L1-negative tumors, squamous histology, liver/bone metastases, low TMB, and STK11 or KRAS variants; these exploratory analyses suggest that other subgroups such as patients with RBM10 variants merit further investigation. These results suggest a potential sensitizing effect from the addition of ceralasertib to durvalumab in post-ICB NSCLC, and our data provide insights into the potential underlying mechanisms of action. Durvalumab–ceralasertib treatment induces systemic effects that associate with immune and inflammatory activities; thus, we hypothesize that these dynamic biomarkers could impact TME immunomodulation and thereby prime enhanced immune-mediated killing of TCs. Ongoing studies are currently investigating baseline and on-treatment tumor biopsies to further elucidate our mechanistic understanding of ceralasertib-mediated immune changes.
Durvalumab–ceralasertib demonstrated a generally acceptable safety profile and was broadly tolerable, with a low rate of treatment discontinuation due to TRAEs. Common TRAEs reflected those seen in previous studies64,65, with no new safety signals. Overall rates of TRAEs, grade ≥3 TRAEs and serious TRAEs were similar or numerically lower with durvalumab–ceralasertib compared with the other regimens, despite a >3-month longer mean duration of treatment. However, as with the efficacy data, these findings should be interpreted in the context of the limited follow-up in a number of patients receiving durvalumab–ceralasertib at data cutoff.
The heterogeneous nature of the NSCLC patient population in this setting highlights one of the benefits of platform or umbrella study designs such as in HUDSON, COAST45, NeoCOAST66 and Lung-MAP67. Modular designs provide the ability to evaluate multiple combinations within a specific treatment setting simultaneously. Additionally, in the context of investigating molecularly targeted treatment, such study designs can encompass patients with a range of different targetable aberrations, potentially resulting in an increased proportion of screened patients meeting specific cohort eligibility requirements, in contrast to single-arm phase 2 studies with a single set of eligibility criteria. Furthermore, central molecular screening of fresh biopsy samples, with molecular characterization of tumors, increased the chances of obtaining hypothesis-generating findings for both the biomarker-matched and biomarker-nonmatched primary and acquired resistance populations. However, the use of different NGS and immunohistochemistry assays for allocation of patients to the biomarker-matched or biomarker-nonmatched cohorts, and of archival or fresh biopsies, in HUDSON highlights the need for robust assessment using standardized biomarker testing with a short turnaround time.
HUDSON and platform studies in general also have limitations associated with their design. HUDSON was designed to allow the addition of new modules to adapt to developing understanding of anti-PD-(L)1 resistance mechanisms. Therefore, it is an open-label, nonrandomized study, and so it is not feasible to conduct formal controlled comparisons between treatment regimens, particularly with the limited cohort sizes. Additionally, because treating patients who had progressed on prior ICB with ICB alone was deemed to not be medically appropriate, it was not feasible to directly compare these combination regimens against durvalumab monotherapy, and so the specific benefit of adding each combination partner to durvalumab could not be determined. Comparison of outcomes between cohorts and regimens is further confounded by differences in patient demographics and baseline disease characteristics associated with the different subsets of patients treated within each cohort.
In conclusion, these findings have demonstrated an efficacy signal of interest with durvalumab–ceralasertib in advanced/metastatic NSCLC following prior failure of anti-PD-1/PD-L1 immunotherapy and platinum-doublet therapy. The regimen showed particular efficacy in patients with ATM alterations and in biomarker-nonmatched primary and acquired resistance cohorts across various subgroups recalcitrant to ICB. Our findings have thus resulted in initiation of the phase 3 LATIFY study (NCT05450692), which compares durvalumab–ceralasertib versus docetaxel for the treatment of patients with NSCLC whose disease progressed during or following prior anti-PD-1/PD-L1 therapy and platinum-based chemotherapy. Additionally, HUDSON is ongoing, with continued accrual to biomarker-matched and biomarker-nonmatched cohorts receiving durvalumab–ceralasertib or ceralasertib monotherapy and to cohorts investigating additional treatment combinations of durvalumab plus trastuzumab deruxtecan or cediranib.
Methods
Study design
HUDSON (NCT03334617) is an ongoing open-label, multicenter, umbrella phase 2 study of the anti-PD-L1 human monoclonal antibody durvalumab in combination with novel molecular-targeted anticancer agents in patients who have progressed on an anti-programmed death-1 (PD-1)/PD-L1-containing therapy and been exposed to a platinum-doublet regimen for locally advanced or metastatic NSCLC, either separately or in combination. HUDSON has a modular design, allowing initial assessment of the efficacy, safety and tolerability of multiple treatment combinations tailored by molecular alteration, with the goal of overcoming resistance to PD-(L)1 blockade.
The study is being performed in accordance with consensus ethical principles derived from international guidelines including the Declaration of Helsinki and Council for International Organizations of Medical Sciences International Ethical Guidelines, applicable International Council for Harmonisation Good Clinical Practice Guidelines, all applicable laws and regulations, and the AstraZeneca policy on Bioethics and Human Biological Samples. The study protocol, protocol amendments, informed consent form, investigator’s brochure and other relevant documents were reviewed and approved by the Institutional Review Board/Ethics Committee at each participating center: Austria—Wiener Gesundheitsverbund–Klinik Floridsdorf; Uniklinikum Salzburg; LBI for Lung Health, c/o Klinik Penzing, Wiener Gesundheitsverbund; Universitätsklinik Innsbruck. Canada—Princess Margaret Cancer Centre; William Osler Health System–Brampton Civic Hospital; Alberta Health Services; The Ottawa Hospital; Centre Hospitalier de l'Universite de Montreal. France—CHU Nantes Hôpital Laennec; Institut Gustave Roussy; Institut Bergonié; Hôpital Bichat–Claude-Bernard. Israel—Rabin Medical Center–Beilinson Hospital; Rambam Health Care Campus; Meir Hospital. South Korea—Samsung Medical Center; Seoul National University Hospital; Asan Medical Center. Spain—Hospital Universitario Ramón y Cajal; Hospital Universitario Virgen Macarena. United States—The University of Texas; Johns Hopkins University; UCLA; Washington University in St. Louis School of Medicine; St Joseph Heritage Healthcare; Dana Farber Mass General Brigham Cancer Care; City of Hope National Medical Center; Fox Chase Cancer Center; Sarah Cannon Research Institute at Tennessee Oncology; New York-Presbyterian; University of California San Diego; Sibley Memorial Hospital; Virginia Cancer Specialists; University of Pittsburgh School of Medicine. Protocol sections covering the modules reported in the manuscript are available as outlined in the data availability statement.
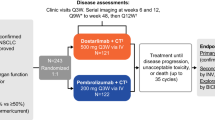
Patients are assigned to either a biomarker-matched group (group A) or a biomarker-nonmatched group (group B) based on tumor molecular profile at screening, as assessed by a consistent genomic testing protocol. Within the biomarker-matched group, patients with tumors with a mutation detected in a homologous recombination repair gene (HRRm) or with detectable aberrations in LKB1 (liver kinase B1; also known as serine threonine kinase 11 (STK11)) were enrolled to separate cohorts to receive durvalumab plus the PARP inhibitor olaparib (module 1); patients with detectable aberrations in ATM (ataxia telangiectasia mutated) on next-generation sequencing (NGS; tissue-based FoundationOne CDx assay68 (Foundation Medicine) or circulating tumor DNA (ctDNA)-based GuardantHealth360 (Guardant Health) assay) or low ATM protein expression on immunohistochemistry (Ventana ATM (Y170) assay) received the ATR protein kinase inhibitor ceralasertib (AZD6738)69 (module 3); and patients with tumors expressing high levels of CD73 received durvalumab plus the anti-CD73 monoclonal antibody oleclumab (MEDI9447) (module 5). Other cohorts within the biomarker-matched group were opened or remain open for patient enrollment but are not described here as data are not sufficiently mature or planned initial accrual has not been completed. Within the biomarker-nonmatched group, patients without prespecified biomarkers are enrolled to cohorts according to whether they had primary or acquired resistance to their prior anti-PD-1/PD-L1 therapy, defined respectively as disease progression within ≤24 weeks or after >24 weeks from the start of treatment, while still on that treatment. At data cutoff for this report (modules 1, 3 and 5: 26 April 2022; module 2: 26 October 2020), data were available from biomarker-nonmatched cohorts treated with durvalumab plus olaparib (module 1), the signal transducer and activator of transcription 3 (STAT3) inhibitor danvatirsen (AZD9150) (module 2), ceralasertib (module 3) and oleclumab (module 5); other cohorts were opened or remain open. For each cohort within the biomarker-matched and biomarker-nonmatched groups, planned enrollment was 20 evaluable patients, with expansion to 40 evaluable patients determined on the basis of efficacy findings. Module 4, investigating durvalumab plus the dual mammalian target of rapamycin complex 1 and 2 (mTORC1/2) inhibitor vistusertib (AZD2014)70 in patients with RPTOR independent companion of MTOR complex 2 (RICTOR) mutations, was halted after one patient had been dosed due to discontinuation of drug development, and is not reported here.
The primary objective is to assess the objective response rate (ORR) for each treatment combination according to Response Evaluation Criteria for Solid Tumours (RECIST) version 1.1 (refs. 71,72). Secondary objectives include the assessment of disease control rate, PFS and OS, as well as the safety and tolerability of each treatment regimen. Exploratory objectives include investigations of cancer-relevant immune status, including biomarker analyses according to specific gene or protein expression profiles (for example, PD-L1), and the usage of subsequent anticancer therapy.
Patients
To be eligible for enrollment, all patients had to be ≥18 years of age and to have histologically or cytologically confirmed metastatic or locally advanced and recurrent NSCLC that was progressing. Patients were required to be eligible for second- or later-line therapy and must have received an anti-PD-1/PD-L1-containing therapy and a platinum-doublet regimen for locally advanced or metastatic NSCLC either separately or in combination. Prior durvalumab was permitted. Patients must have had disease progression on a prior line of anti-PD-1/PD-L1 therapy. Patients needed to be suitable for a new tumor biopsy or to have had a biopsy post-progression on an anti-PD-1/PD-L1 containing therapy within approximately 3 months of screening, and required ≥1 lesion that could be accurately measured predose as ≥10 mm in the longest diameter (except for lymph nodes, for which the requirement was a short axis ≥15 mm) using computed tomography (CT) or magnetic resonance imaging and that was suitable for accurate repeated measurements. Additionally, patients were required to have a body weight of >30 kg, no cancer-associated cachexia, an Eastern Cooperative Oncology Group (ECOG) performance status of 0 to 1, a minimum life expectancy of 12 weeks, and a treatment-free interval of ≥3 weeks from any prior therapy before starting study treatment.
Patients were excluded if their tumors had targetable alterations in EGFR and/or ALK or were known to have targetable alterations in ROS1, BRAF, MET or RET. Patients must not have had toxicity that led to permanent discontinuation of prior anti-PD-1/PD-L1 immunotherapy, or any grade ≥3 immune-related adverse event (AE) or an immune-related neurologic or ocular AE of any grade while receiving prior immunotherapy. All AEs that occurred while receiving prior immunotherapy must have completely resolved or resolved to baseline before screening. Other exclusion criteria were the following: history of active primary immunodeficiency; active or prior documented autoimmune or inflammatory disorders (except vitiligo, alopecia, hypothyroidism that was stable on hormone replacement therapy, any chronic skin condition not requiring systemic therapy, or celiac disease controlled by diet alone); a history of allogenic organ transplantation; any uncontrolled intercurrent illness; spinal cord compression or symptomatic brain metastases; active infection; history of another primary malignancy (except malignancies treated with curative intent and with no known active disease ≥2 years before the first dose of study drug and of low potential risk for recurrence, adequately treated nonmelanoma skin cancer or lentigo maligna without evidence of disease, adequately treated carcinoma in situ without evidence of disease, and localized noninvasive primary under surveillance); small-cell lung cancer; presence of prespecified cardiac criteria; and inadequate bone marrow reserve or organ function.
Patients could not be receiving any concurrent chemotherapy, immunotherapy, biologic or hormonal therapy for cancer treatment, but use of hormones for non-cancer-related conditions and local treatment of isolated lesions, excluding target lesions, for palliative intent were acceptable. Current or prior use of immunosuppressive medication within 14 days before the first dose of durvalumab was not permitted (except for intranasal, inhaled, topical steroids or local steroid injections; systemic corticosteroids at doses not exceeding 10 mg per day of prednisone or its equivalent; steroids as premedication for hypersensitivity reactions). Patients were not allowed to have received a live attenuated vaccine within 30 days before the first dose of study drug, although authorized/approved coronavirus disease 2019 vaccines were permitted, with the recommendation to avoid administration for 72 h before the first dose of study drug.
All patients provided written informed consent. Prescreening consent was obtained for access to preexisting molecular information (if available), testing of an archival tumor sample (if available) and/or plasma ctDNA, and provision of a new tumor biopsy sample and blood sample for ctDNA testing. Following prescreening, and a valid molecular analysis result, patients were assigned to, and invited to consent to, a specific treatment cohort.
Treatment and assessments
All patients received durvalumab 1,500 mg via intravenous infusion once every 4 weeks. Patients also received: olaparib orally at a dose of 300 mg twice daily (module 1 cohorts); danvatirsen intravenously at a dose of 200 mg every other day for a 1-week lead-in period before starting durvalumab and then weekly in combination with durvalumab (module 2 cohorts); ceralasertib orally at a dose of 240 mg twice daily on days 1–7 of a 1-week lead-in period before starting durvalumab and then on days 22–28 of each 4-week cycle in combination with durvalumab (module 3 cohorts); or oleclumab intravenously at a dose of 3,000 mg once every 2 weeks in cycles 1–2 and then once every 4 weeks from cycle 3 onwards (fixed dosing for patients with body weight >30 kg) (module 5 cohorts).
Tumors were assessed using contrast enhanced CT (or magnetic resonance imaging if CT was contraindicated) scans of the chest, abdomen and pelvis (including liver and adrenal glands) at baseline, every 6 weeks (±1 week) for the first 24 weeks from the start of combination therapy, and every 8 weeks (±1 week) thereafter until disease progression (confirmed with a subsequent scan). Patients who, in the opinion of the investigator, were still receiving clinical benefit from treatment, were permitted to continue treatment following confirmed radiological disease progression per RECIST v1.1 (refs. 71,72). Scheduled radiological scans were continued for these patients while they were still receiving study treatment. Objective tumor responses were assessed and categorized programmatically per RECIST v1.1 (refs. 71,72). Survival status was recorded every 3 months (±1 week) after a safety follow-up visit (90 days after study drug discontinuation) and until the earlier of: 12 months after the last patient had discontinued treatment in the last cohort within a module; or 75% of patients had died in all cohorts within a module. Safety was assessed and AEs recorded throughout the treatment period and the safety follow-up (90 days after the discontinuation of all study drugs or until initiation of another therapy, unless the investigator assesses that the event occurring within 90 days after last dose of study treatment but after the initiation of another therapy is related to the study treatment). Severity of AEs was assessed per the National Cancer Institute’s Common Terminology Criteria for Adverse Events version 4.03. Causal relationship between study drug and each AE was assessed by the investigators, by answering ‘yes’ or ‘no’ to the question ‘Do you consider that there is a reasonable possibility that the event may have been caused by the study drug?’.
Biomarker analyses
A collection of tumor samples, including both resections (archival) and post-anti-PD-1/PD-L1 progression biopsies (new), was assembled from patients during screening. Each sample was sequenced using the Foundation Medicine NGS FoundationOneCDx assay68. The assay detects mutations and copy number variations in 324 cancer-related genes and selected rearrangements. The assay also provides information on microsatellite instability and TMB. Detailed information on the assay and the variant calling pipeline are available at ref. 73. TMB is reported as low (<10 mutations Mb−1) or high (≥10 mutations Mb−1). Mutation profiles included co-occurring and mutually exclusive gene alterations. For each gene, substitutions, short insertions and deletions, rearrangements and copy number changes of known or likely functional relevance detected using the assay were included. Additionally, tumor PD-L1 expression was assessed locally using PD-L1 immunohistochemistry assays. PD-L1 expression was evaluated in TCs and considered positive when expressed in ≥1% of neoplastic cells.
RNA extraction and whole transcriptome library preparation
Bulk-RNA sequencing was conducted on suitable fresh biopsies acquired from a subset of patients with evaluable prescreening tumor biopsies, before patient allocation to a HUDSON treatment module. Fresh frozen core needle biopsy samples were divided (where possible) while frozen on dry ice, with one portion consumed for total RNA extraction. Total RNA was extracted using Qiagen RNeasy Mini kit (Qiagen, cat. no. 74104) with on-column DNase digestion (cat. no. 79254) and eluted in 30 ml nuclease-free water. RNA concentration, RNA integrity number and %DV200 (percentage of RNA fragments >200 nucleotides) were determined using Agilent RNA ScreenTapes or Agilent High Sensitivity RNA ScreenTapes via Agilent TapeStation. Total RNA was arrayed on a 96-well polymerase chain reaction plate, and whole transcriptome libraries were generated using KAPA RNA Hyper Prep Kit with RiboErase (HMR) Globin (Roche cat. no. KK8563), in accordance with the manufacturer’s protocol. Library concentrations were determined using Agilent D1000 ScreenTapes on Agilent TapeStation. Whole transcriptome sequencing libraries were sequenced with the Illumina NovaSeq 6000 kit v1.5 300 cycles (150 × 2).
Transcript-per-million (TPM)-normalized bulk-RNA sequencing data were available for 49 prescreening samples from 49 unique patients. The TPM matrix was used as input for single-sample gene set enrichment analysis (ssGSEA) as implemented in GSVA R package version 1.42.0 to compute enrichment scores (ES) for the 18-gene TIS47. ssGSEA ES were compared between patients with immunotherapy-refractory/relapsed NSCLC, including recalcitrant subgroups, using the two-sided Wilcoxon rank-sum test. Specifically, TIS ssGSEA ES was compared between patients with primary (progression within 24 weeks) or acquired (progression after 24 weeks) resistance to prior ICB therapy, patients with PD-L1-positive (≥1% positive TCs based on immunohistochemical staining) or PD-L1-negative (<1% TCs) tumors, patients with or without bone and/or liver metastases, and patients with adenocarcinoma or squamous cell tumors. In addition, comprehensive genomic profiling using targeted next-generation sequencing was performed on archived or baseline tissue samples using the FoundationOne CDx assay68. Detailed information on the assay and the variant calling pipeline is available at ref. 73. Based on the mutational status, TIS ssGSEA ES were compared between patients with STK11-altered or STK11-wild-type tumors, and patients with TMB-low (<10 mutations Mb−1) or TMB-high (≥10 mutations Mb−1) tumors. The TPM matrix was z-scored gene-wise to display the expression level of the 18 genes composing the TIS as heat maps. It is important to note that analyses were not restricted to specific biopsy sites because of the limited numbers of prescreening biopsies assessed using bulk-RNA sequencing.
RNA extraction and NanoString gene expression
Total RNA was extracted from pre- and posttreatment blood samples using the PAXgene Blood RNA Kit (PreAnalytiX GmbH) per the manufacturer′s recommended protocol. RNA concentration and quality were assessed on the TapeStation 2200 System (Agilent). nCounter gene expression assays (NanoString Technologies) were performed on patients’ blood RNA samples using the 770-gene human PanCancer Immune Profiling panel74 (NanoString Technologies) and a custom 30-gene spike-in panel, per NanoString′s recommended protocol. The custom spike-in panel target genes included APOBEC3B, B2M, CALB2, CD69, CGAS, CMPK2, DHX58, EGFR, GBP1, GBP4, GBP5, GDF15, HAP1, HOPX, IFNG, IL12A, KEAP1, NKG7, OASL, PF4, PROM1, RGS16, RSAD2, SAP30, SLAMF8, STING1, STK11, TREX1, TSLP and VTCN1. Raw count data were preprocessed using background subtraction and housekeeping gene normalization functions in nSolver 4.0 (NanoString Technologies). The data that passed quality control using nSolver’s default quality control parameters were then analyzed using a linear mixed effects model.
TCR sequencing and analysis
Deep sequencing of the CDR3 regions of TCR-β genes was performed by Adaptive Biotechnologies using the immunoSEQ assay on genomic DNA purified with the Qiagen DNeasy Blood extraction kit from total peripheral blood mononuclear cells obtained at each study visit. Only productive (in-frame) TCR sequences were considered in the analyses75,76. To identify expanded and contracted T cell clones between samples from an individual patient at different visits, we used the Fisher exact test to compute a P value for each TCR clonotype across the two samples, against the null hypothesis that the population abundance of the clone is identical in the two samples. We corrected for multiple testing to control the false discovery rate using the Benjamini–Hochberg procedure and employed a significance threshold of 0.01 on adjusted P values. Expanded clones not observed at baseline and detected above a threshold of five reads were classified as newly detected expanded clones. We calculated Simpson’s clonality (as 1 − Pielou’s evenness), which normalizes sampling depth to compare TCR repertoires between samples. To measure the stability of the clonotypes in the circulating repertoire across visits, we used the Morisita–Horn similarity index, which accounts for the number of common clonotypes and the distribution of clonotype sizes and is most sensitive to the clone sizes of the dominant clonotypes75,76. Wilcoxon paired signed-rank tests were used to compare groups of matched samples.
Statistical analysis
Initial sample size for each cohort was to be 20 evaluable patients, with possible expansion based on observation of an efficacy signal per the primary endpoint of ORR. Evaluations were to be carried out after the ~20th evaluable patient in a cohort, or the final patient dosed in a cohort if enrollment ended early, had had the opportunity for two on-treatment response assessments per RECIST or had discontinued or withdrawn from treatment. A statistical framework was built around a prespecified ORR to inform the Sponsor if an efficacy signal had been observed and a recommendation to expand should be made. Decisions regarding stopping recruitment in specific cohorts were at the discretion of the Sponsor and were based on emerging efficacy, safety and tolerability data.
PFS was defined as the time from start of treatment until the date of objective disease progression according to RECIST 1.1 or death (by any cause in the absence of progression) regardless of whether the patient withdrew from therapy or received another anticancer therapy before progression. Patients who had not progressed or died at the time of analysis were censored at the time of the latest date of assessment from their last evaluable RECIST 1.1 assessment. However, if a patient progressed or died after two or more missed visits, the patient was censored at the time of the latest evaluable RECIST 1.1 assessment before the two missed visits. OS was defined as the time from the start of treatment until death due to any cause. Any patient not known to have died at the time of analysis was censored on the basis of the last recorded date on which the patient was known to be alive. PFS and OS were evaluated using Kaplan–Meier methodology. Analyses of clinical data were conducted using SAS v9.3.
Data cutoff for modules 1, 3 and 5 was 26 April 2022. Due to the nature of this platform study, which enrolled different cohorts at different time periods, data cutoff for module 2 was 26 October 2020.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The relevant sections of the study protocol, plus data underlying the findings described in this paper, may be obtained in accordance with AstraZeneca’s data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. Data for studies directly listed on Vivli can be requested through Vivli at www.vivli.org. Data for studies not listed on Vivli could be requested through Vivli at https://vivli.org/members/enquiries-about-studies-not-listed-on-the-vivli-platform/. AstraZeneca’s Vivli member page is also available outlining further details (https://vivli.org/ourmember/astrazeneca/).
References
NCCN clinical practice guidelines in oncology: non-small cell lung cancer, version 3.2022. National Comprehensive Cancer Network https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1450 (2022).
Lamberti, G. et al. Beyond EGFR, ALK and ROS1: current evidence and future perspectives on newly targetable oncogenic drivers in lung adenocarcinoma. Crit. Rev. Oncol. Hematol. 156, 103119 (2020).
Sholl, L. M. et al. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: the Lung Cancer Mutation Consortium experience. J. Thorac. Oncol. 10, 768–777 (2015).
Lam, V. K. et al. Targeted tissue and cell-free tumor DNA sequencing of advanced lung squamous-cell carcinoma reveals clinically significant prevalence of actionable alterations. Clin. Lung Cancer 20, 30–36 e33 (2019).
Brahmer, J. et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 373, 123–135 (2015).
Herbst, R. S. et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387, 1540–1550 (2016).
Reck, M. et al. Five-year outcomes with pembrolizumab versus chemotherapy for metastatic non-small-cell lung cancer with PD-L1 tumor proportion score ≥ 50. J. Clin. Oncol. 39, 2339–2349 (2021).
Rittmeyer, A. et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 389, 255–265 (2017).
Borghaei, H. et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 373, 1627–1639 (2015).
Gandhi, L. et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 378, 2078–2092 (2018).
West, H. et al. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 20, 924–937 (2019).
Paz-Ares, L. et al. First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): an international, randomised, open-label, phase 3 trial. Lancet Oncol. 22, 198–211 (2021).
Boyero, L. et al. Primary and acquired resistance to immunotherapy in lung cancer: unveiling the mechanisms underlying of immune checkpoint blockade therapy. Cancers 12, 3729 (2020).
Schoenfeld, A. J. & Hellmann, M. D. Acquired resistance to immune checkpoint inhibitors. Cancer Cell 37, 443–455 (2020).
Yuan, Y., Adam, A., Zhao, C. & Chen, H. Recent advancements in the mechanisms underlying resistance to PD-1/PD-L1 blockade immunotherapy. Cancers 13, 663 (2021).
Walsh, R. J. & Soo, R. A. Resistance to immune checkpoint inhibitors in non-small cell lung cancer: biomarkers and therapeutic strategies. Ther. Adv. Med Oncol. 12, 1758835920937902 (2020).
Aguilar, E. J. et al. Outcomes to first-line pembrolizumab in patients with non-small-cell lung cancer and very high PD-L1 expression. Ann. Oncol. 30, 1653–1659 (2019).
Hellmann, M. D. et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 378, 2093–2104 (2018).
Kluger, H. M. et al. Defining tumor resistance to PD-1 pathway blockade: recommendations from the first meeting of the SITC Immunotherapy Resistance Taskforce. J. Immunother. Cancer 8, e000398 (2020).
Lee, Y. J., Lee, J. B., Ha, S. J. & Kim, H. R. Clinical perspectives to overcome acquired resistance to anti-programmed death-1 and anti-programmed death ligand-1 therapy in non-small cell lung cancer. Mol. Cells 44, 363–373 (2021).
Schoenfeld, A. J. et al. Clinical definition of acquired resistance to immunotherapy in patients with metastatic non-small-cell lung cancer. Ann. Oncol. 32, 1597–1607 (2021).
Koyama, S. et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res. 76, 999–1008 (2016).
Rizvi, H. et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 36, 633–641 (2018).
Skoulidis, F. et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 8, 822–835 (2018).
Skoulidis, F. et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 5, 860–877 (2015).
Pore, N. et al. Resistance to durvalumab and durvalumab plus tremelimumab is associated with functional STK11 mutations in patients with non-small cell lung cancer and is reversed by STAT3 knockdown. Cancer Discov. 11, 2828–2845 (2021).
Papillon-Cavanagh, S., Doshi, P., Dobrin, R., Szustakowski, J. & Walsh, A. M. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open 5, e000706 (2020).
Kerr, K. M. et al. The evolving landscape of biomarker testing for non-small cell lung cancer in Europe. Lung Cancer 154, 161–175 (2021).
Bodor, J. N., Boumber, Y. & Borghaei, H. Biomarkers for immune checkpoint inhibition in non-small cell lung cancer (NSCLC). Cancer 126, 260–270 (2020).
Govindan, R. et al. Society for Immunotherapy of Cancer (SITC) clinical practice guideline on immunotherapy for the treatment of lung cancer and mesothelioma. J. Immunother. Cancer 10, e003956 (2022).
Keenan, T. E., Burke, K. P. & Van Allen, E. M. Genomic correlates of response to immune checkpoint blockade. Nat. Med. 25, 389–402 (2019).
Mouw, K. W., Goldberg, M. S., Konstantinopoulos, P. A. & D’Andrea, A. D. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 7, 675–693 (2017).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Huang, J. L., Chang, Y. T., Hong, Z. Y. & Lin, C. S. Targeting DNA damage response and immune checkpoint for anticancer therapy. Int. J. Mol. Sci. 23, 3238 (2022).
Germano, G. et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552, 116–120 (2017).
Linnebacher, M., Wienck, A., Boeck, I. & Klar, E. Identification of an MSI-H tumor-specific cytotoxic T cell epitope generated by the (−1) frame of U79260(FTO). J. Biomed. Biotechnol. 2010, 841451 (2010).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Rouleau, M., Patel, A., Hendzel, M. J., Kaufmann, S. H. & Poirier, G. G. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 10, 293–301 (2010).
Fong, P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 (2009).
Weber, A. M. & Ryan, A. J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 149, 124–138 (2015).
Yap, T. A. et al. Ceralasertib (AZD6738), an oral ATR kinase inhibitor, in combination with carboplatin in patients with advanced solid tumors: a phase I study. Clin. Cancer Res. 27, 5213–5224 (2021).
Rebe, C. & Ghiringhelli, F. STAT3, a master regulator of anti-tumor immune response. Cancers 11, 1280 (2019).
Cohen, E. E. W. et al. A phase Ib/II study (SCORES) of durvalumab (D) plus danvatirsen (DAN; AZD9150) or AZD5069 (CX2i) in advanced solid malignancies and recurrent/metastatic head and neck squamous cell carcinoma (RM-HNSCC): updated results. Ann. Oncol. 29, viii372 (2018).
Allard, B., Allard, D., Buisseret, L. & Stagg, J. The adenosine pathway in immuno-oncology. Nat. Rev. Clin. Oncol. 17, 611–629 (2020).
Herbst, R. S. et al. COAST: an open-label, phase II, multidrug platform study of durvalumab alone or in combination with oleclumab or monalizumab in patients with unresectable, stage III non-small-cell lung cancer. J. Clin. Oncol. 40, 3383–3393 (2022).
Long, L. L. et al. PARP inhibition induces synthetic lethality and adaptive immunity in LKB1-mutant lung cancer. Cancer Res. 83, 568–581 (2023).
Danaher, P. et al. Pan-cancer adaptive immune resistance as defined by the tumor inflammation signature (TIS): results from The Cancer Genome Atlas (TCGA). J. Immunother. Cancer 6, 63 (2018).
Ayers, M. et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017).
Damotte, D. et al. The tumor inflammation signature (TIS) is associated with anti-PD-1 treatment benefit in the CERTIM pan-cancer cohort. J. Transl. Med 17, 357 (2019).
Gutierrez, M. et al. Biomarker-directed, pembrolizumab-based combination therapy in non-small cell lung cancer: phase 2 KEYNOTE-495/KeyImPaCT trial interim results. Nat. Med. 29, 1718–1727 (2023).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Danaher, P. et al. Gene expression markers of tumor infiltrating leukocytes. J. Immunother. Cancer 5, 18 (2017).
Nadal, E. et al. Two patients with advanced-stage lung adenocarcinoma with radiologic complete response to nivolumab treatment harboring an STK11/LKB1 mutation. JCO Precis. Oncol. 4, 1239–1245 (2020).
Neal, J. et al. CONTACT-01: efficacy and safety from a phase III study of atezolizumab (atezo) + cabozantinib (cabo) vs docetaxel (doc) monotherapy in patients (pts) with metastatic NSCLC (mNSCLC) previously treated with checkpoint inhibitors and chemotherapy. J. Thorac. Oncol. 18, S35–S88 (2023). abstract 60.
Besse, B. et al. Randomized open-label controlled study of cancer vaccine OSE2101 versus chemotherapy in HLA-A2-positive patients with advanced non-small-cell lung cancer with resistance to immunotherapy: ATALANTE-1. Ann. Oncol. 34, 920–933 (2023).
de Langen, A. J. et al. Sotorasib versus docetaxel for previously treated non-small-cell lung cancer with KRAS(G12C) mutation: a randomised, open-label, phase 3 trial. Lancet 401, 733–746 (2023).
Mazieres, J. et al. Atezolizumab versus docetaxel in pretreated patients with NSCLC: final results from the randomized phase 2 POPLAR and phase 3 OAK clinical trials. J. Thorac. Oncol. 16, 140–150 (2021).
Lu, S. et al. Nivolumab versus docetaxel in a predominantly Chinese patient population with previously treated advanced non-small cell lung cancer: 2-year follow-up from a randomized, open-label, phase 3 study (CheckMate 078). Lung Cancer 152, 7–14 (2021).
Borghaei, H. et al. Five-year outcomes from the randomized, phase III trials CheckMate 017 and 057: nivolumab versus docetaxel in previously treated non-small-cell lung cancer. J. Clin. Oncol. 39, 723–733 (2021).
Herbst, R. S. et al. Five year survival update from KEYNOTE-010: pembrolizumab versus docetaxel for previously treated, programmed death-ligand 1-positive advanced NSCLC. J. Thorac. Oncol. 16, 1718–1732 (2021).
Park, K. et al. Avelumab versus docetaxel in patients with platinum-treated advanced NSCLC: 2-year follow-up from the JAVELIN Lung 200 Phase 3 Trial. J. Thorac. Oncol. 16, 1369–1378 (2021).
Billan, S., Kaidar-Person, O. & Gil, Z. Treatment after progression in the era of immunotherapy. Lancet Oncol. 21, e463–e476 (2020).
Goeman, F. et al. Mutations in the KEAP1–NFE2L2 pathway define a molecular subset of rapidly progressing lung adenocarcinoma. J. Thorac. Oncol. 14, 1924–1934 (2019).
Kim, R. et al. Phase II study of ceralasertib (AZD6738) in combination with durvalumab in patients with advanced/metastatic melanoma who have failed prior anti-PD-1 therapy. Ann. Oncol. 33, 193–203 (2022).
Rizvi, N. A. et al. Durvalumab with or without tremelimumab vs standard chemotherapy in first-line treatment of metastatic non-small cell lung cancer: the MYSTIC Phase 3 Randomized Clinical Trial. JAMA Oncol. 6, 661–674 (2020).
Guisier, F. et al. NeoCOAST-2: a phase 2 study of neoadjuvant durvalumab plus novel immunotherapies (IO) and chemotherapy (CT) or MEDI5752 (volrustomig) plus CT, followed by surgery and adjuvant durvalumab plus novel IO or volrustomig alone in patients with resectable non-small-cell lung cancer (NSCLC). J. Clin. Oncol. 41, TPS8604 (2023).
Redman, M. W. et al. Biomarker-driven therapies for previously treated squamous non-small-cell lung cancer (Lung-MAP SWOG S1400): a biomarker-driven master protocol. Lancet Oncol. 21, 1589–1601 (2020).
Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013).
Foote, K. M. et al. Discovery and characterization of AZD6738, a potent inhibitor of ataxia telangiectasia mutated and Rad3 related (ATR) kinase with application as an anticancer agent. J. Med. Chem. 61, 9889–9907 (2018).
Morscher, R. J. et al. First-in-child phase I/II study of the dual mTORC1/2 inhibitor vistusertib (AZD2014) as monotherapy and in combination with topotecan-temozolomide in children with advanced malignancies: arms E and F of the AcSe-ESMART trial. Eur. J. Cancer 157, 268–277 (2021).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Schwartz, L. H. et al. RECIST 1.1—update and clarification: from the RECIST committee. Eur. J. Cancer 62, 132–137 (2016).
FoundationOne CDx technical information. Foundation Medicine https://www.rochefoundationmedicine.com/f1cdxtech (2023).
Cesano, A. nCounter((R)) PanCancer immune profiling panel. J. Immunother. Cancer 3, 42 (2015).
Robins, H. S. et al. Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood 114, 4099–4107 (2009).
DeWitt, W. S. et al. Dynamics of the cytotoxic T cell response to a model of acute viral infection. J. Virol. 89, 4517–4526 (2015).
Acknowledgements
Medical writing support for the development of this manuscript, under the direction of the authors, was provided by S. Hill, of Ashfield MedComms, an Inizio company, and was funded by AstraZeneca.
Author information
Authors and Affiliations
Contributions
B.B., S.I., S.T.B., J.C. and J.V.H. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study conception and design: B.B., K.P., P.M.F., M.J.H., M.M.A., G.G., R.H., H.J.A., J.C.B., M.D., P.J.J., S.I., S.T.B., J.C. and J.V.H. Acquisition of data: B.B., E.P.-T., K.P., S.H., P.M.F., M.M.A., M.T., G.G., P.W.-P., F.A.S., M.F., P.C., Q.S.C.C., S.-W.K., D.M., M.L.J., S.C., M.T.M., S.K., D.L.R., E.D., S.I., J.C. and J.V.H. Analysis of data: B.B., K.P., S.H., P.M.F., M.M.A., M.T., G.G., P.W.-P., F.A.S., Q.S.C.C., S.-W.K., B.A., R.H., S.K., A.R., D.L.R., M.R.K., J.P.C., S.I., J.C. and J.V.H. Interpretation of data: B.B., K.P., S.H., P.M.F., M.M.A., M.T., Q.S.C.C., D.V., B.A., R.H., S.K., A.R., J.P.C., J.C.B., E.D., R.K., M.D., P.J.J., S.I., S.T.B., J.C. and J.V.H. Drafting of the paper: B.A., S.I., S.T.B., J.C. and J.V.H. Critical revision of the paper for important intellectual content: all authors. Statistical analysis: R.H., S.K., A.R., M.R.K., J.P.C. and S.I. Approval of final version of the paper for submission: all authors.
Corresponding author
Ethics declarations
Competing interests
B.B. has received grants or funds from Abbvie, Amgen, AstraZeneca, Chugai Pharmaceutical, Daiichi Sankyo, Ellipse Pharma, EISAI, Genmab, Genzyme Corporation, Hedera Dx, Inivata, IPSEN, Janssen, MSD, Pharmamar, Roche-Genentech, Sanofi, Socar Research, Tahio Oncology and Turning Point Therapeutics. E.P.-T. has participated in advisory councils or committees for AstraZeneca, Bristol Myers Squibb, Takeda and Sanofi. K.P. has participated in an advisory council or committee (steering committee) for and received grants or funds (out of this study) from AstraZeneca. S.H. has participated in advisory boards for MSD, Roche and AstraZeneca, and has received institutional grants or funds from AstraZeneca, GSK, Chiesi and Menarini Pharma. P.M.F. has received consulting fees from AstraZeneca, Bristol Myers Squibb, Novartis, Merck, Genentech, G1, F Star, Regeneron, Janssen, Sanofi, Amgen, Fosun, Teva, Synthekine, Flame, Iteos and Tavotek, and grants or funds from AstraZeneca, Bristol Myers Squibb, Regeneron, Novartis and BioNTech. M.J.H. has participated in advisory councils or committees for Bristol Myers Squibb, AstraZeneca, MSD, Lilly and Roche. M.M.A. has performed consulting or advisory roles with Merck, Pfizer, Bristol Myers Squibb, Foundation Medicine, Novartis, Gritstone Oncology, Mirati Therapeutics, EMD Serono, AstraZeneca, Instil Bio, Regeneron and Janssen, and has received institutional research funding from Genentech/Roche, Lilly, AstraZeneca, Bristol Myers Squibb, Amgen and Affini-T Therapeutics. M.T. has participated in advisory councils or committees for and received honoraria and consulting fees from AstraZeneca, Roche, Bristol Myers Squibb, MSD, Novartis, Sanofi, Takeda, Pfizer, Merck, Daiichi Sankyo, Janssen, Lilly and Boehringer Ingelheim, and has received grants or funds from AstraZeneca, Roche and Takeda. G.G. has received honoraria from AstraZeneca. P.W.-P. has participated in advisory councils or committees for AstraZeneca, Bristol Myers Squibb, Sanofi, Roche, Novartis, Jazz Pharmaceuticals and Pfizer, and has received honoraria from Bayer, Sanofi and Merck. F.A.S. has participated in advisory boards for and received honoraria and consulting fees from AstraZeneca, and their institution has received clinical trial payments from AstraZeneca. M.F. has participated in advisory councils or committees for AstraZeneca, Bristol Myers Squibb and Takeda, has received honoraria from AstraZeneca and Bristol Myers Squibb, and has received grants or funds from AstraZeneca. P.C. has participated in advisory councils or committees for AstraZeneca and Novartis, received honoraria from AstraZeneca, Janssen, Eli Lilly, GSK, Merck, Sanofi and Amgen, and received consulting fees from AstraZeneca, Bristol Myers Squibb, Roche, Amgen, Pfizer, Novartis and Merck. Q.S.C.C. has received honoraria and consulting fees from Abbvie, Amgen, AnHeart, AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Esperas Pharma, Eisai, Jazz Pharmaceutical, Janssen/Johnson and Johnson, Merck, Novartis, Ocellaris, Pfizer, Roche and Takeda. S.-W.K. has no conflicts of interest to declare. D.M. has received consulting fees from Abbvie, Arcus, Mirati and Lilly, and grants or funds from Merck, AstraZeneca, Incyte, Bristol Myers Squibb, Surface, Epicentrx, Boehringer Ingelheim, Y-mabs, Roche, Lilly, Pfizer and Novartis. M.L.J. has received institutional research funding from AbbVie, Acerta, Adaptimmune, Amgen, Apexigen, Arcus Biosciences, Array BioPharma, Artios Pharma, AstraZeneca, Atreca, BeiGene, BerGenBio, BioAtla, Black Diamond, Boehringer Ingelheim, Bristol Myers Squibb, Calithera Biosciences, Carisma Therapeutics, Checkpoint Therapeutics, City of Hope National Medical Center, Corvus Pharmaceuticals, Curis, CytomX, Daiichi Sankyo, Dracen Pharmaceuticals, Dynavax, Lilly, Elicio Therapeutics, EMD Serono, EQRx, Erasca, Exelixis, Fate Therapeutics, Genentech/Roche, Genmab, Genocea Biosciences, GlaxoSmithKline, Gritstone Oncology, Guardant Health, Harpoon, Helsinn Healthcare SA, Hengrui Therapeutics, Hutchison MediPharma, IDEAYA Biosciences, IGM Biosciences, Immunitas Therapeutics, Immunocore, Incyte, Janssen, Jounce Therapeutics, Kadmon Pharmaceuticals, Kartos Therapeutics, Loxo Oncology, Lycera, Memorial Sloan-Kettering, Merck, Merus, Mirati Therapeutics, Mythic Therapeutics, NeoImmune Tech, Neovia Oncology, Novartis, Numab Therapeutics, Nuvalent, OncoMed Pharmaceuticals, Palleon Pharmaceuticals, Pfizer, PMV Pharmaceuticals, Rain Therapeutics, RasCal Therapeutics, Regeneron Pharmaceuticals, Relay Therapeutics, Revolution Medicines, Ribon Therapeutics, Rubius Therapeutics, Sanofi, Seven and Eight Biopharmaceuticals/Birdie Biopharmaceuticals, Shattuck Labs, Silicon Therapeutics, Stem CentRx, Syndax Pharmaceuticals, Takeda Pharmaceuticals, Tarveda, TCR2 Therapeutics, Tempest Therapeutics, Tizona Therapeutics, TMUNITY Therapeutics, Turning Point Therapeutics, University of Michigan, Vyriad, WindMIL Therapeutics and Y-mAbs Therapeutics; and has received institutional payments for consulting or advisory roles for AbbVie, Amgen, Arcus Biosciences, Arrivent, Astellas, AstraZeneca, Black Diamond, Boehringer Ingelheim, Calithera Biosciences, Daiichi Sankyo, EcoR1, Genentech/Roche, Genmab, Genocea Biosciences, Gilead Sciences, GlaxoSmithKline, Gritstone Oncology, Ideaya Biosciences, Immunocore, iTeos, Janssen, Jazz Pharmaceuticals, Merck, Mirati Therapeutics, Molecular Axiom, Normunity, Novartis, Oncorus, Pyramid Biosciences, Regeneron Pharmaceuticals, Revolution Medicines, Sanofi-Aventis, SeaGen, Synthekine, Takeda Pharmaceuticals, Turning Point Therapeutics and VBL Therapeutics. S.C. has participated in advisory councils or committees for AstraZeneca, Bristol Myers Squibb, MSD and Sanofi. D.-W.K. has received research funding to their institution from Alpha Biopharma, Amgen, AstraZeneca/Medimmune, Boehringer Ingelheim, Bridge BioTherapeutics, Chong Keun Dang, Daiichi Sankyo, GSK, Hanmi, InnoN, Janssen, Merck, Merus, Mirati Therapeutics, MSD, Novartis, ONO Pharmaceutical, Pfizer, Roche/Genentech, Takeda, TP Therapeutics, Xcovery and Yuhan, and has received medical writing assistance from Amgen, AstraZeneca, Boehringer Ingelheim, Bridge BioTherapeutics, Chong Keun Dang, Daiichi Sankyo, GSK, Janssen, Merus, Mirati Therapeutics, MSD, Meck, Novartis, Pfizer, Roche, Takeda and Yuhan. M.T.M. has participated in advisory councils or committees for MSD, Takeda, Bayer, Amgen and Novartis, has received honoraria from Bristol Myers Squibb, MSD, Takeda, AstraZeneca, Roche and AbbVie, has received consulting fees from MSD and AstraZeneca, and has received grants or funds from AstraZeneca. D.V. has received honoraria from Bristol Myers Squibb, AstraZeneca, Pfizer, Novartis, Roche and Daichii. B.A., R.H., H.J.A., S.K., A.R., D.L.R., M.R.K., J.P.C., J.C.B., E.D., R.K., M.D., P.J.J., S.I., S.T.B. and J.C. are employees of and may own stock/shares in AstraZeneca. D.L.R. owns stocks or shares in Myriad Genetics. J.V.H. has participated in advisory councils or committees for AstraZeneca, Bristol Myers Squibb, Genentech/Roche, EMD Serono, GlaxoSmithKline, Hengrui Therapeutics, Eli Lilly, Novartis, Spectrum, Sanofi, Takeda, Mirati Therapeutics, Janssen Global Services and Leads Biolabs, has received honoraria from AstraZeneca, Bristol Myers Squibb, Genentech/Roche, EMD Serono, GlaxoSmithKline, Hengrui Therapeutics, Eli Lilly, Novartis, Spectrum, Sanofi, Takeda, Mirati Therapeutics, Janssen Global Services, Leads Biolabs and Boehringer Ingelheim, has received grants or funds from AstraZeneca, Spectrum, Boehringer Ingelheim and Takeda, and has received licensing fees from Spectrum.
Peer review
Peer review information
Nature Medicine thanks Christopher Cabanski and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ulrike Harjes, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 PFS and OS by treatment cohort.
PFS and OS with a, b, durvalumab-ceralasertib (median OS not yet mature in ATM biomarker-matched cohort), c, d, durvalumab-olaparib, e, f, durvalumab-danvatirsen, g, h, durvalumab-oleclumab.
Extended Data Fig. 2 Gene expression profiling in subgroups with known association with poor immunotherapy response among patients who received durvalumab plus olaparib, danvatirsen or oleclumab.
Gene-wise transcript-per-million (TPM) z-score heatmaps for the 18 genes comprising the tumour inflammation signature (TIS) in tumour samples from patients with a, primary or acquired resistance to prior immune checkpoint blockade therapy (n = 18, n = 31; overall prevalence: n = 81 [43.1%], n = 107 [56.9%]), b, PD-L1-positive or PD-L1-negative tumours (n = 16, n = 18; overall prevalence: n = 73 [58.9%], n = 51 [41.1%]), c, adenocarcinoma or squamous cell histology (n = 38, n = 6; overall prevalence: n = 130 [75.1%], n = 43 [24.9%]), d, with or without bone/liver metastases (n = 23, n = 26; overall prevalence: n = 76 [40.4%], n = 112 [59.6%]), e, high or low TMB (n = 16, n = 18; overall prevalence: n = 60 [41.1%], n = 86 [58.9%]) and f, STK11-altered or STK11-wild-type tumours (n = 5, n = 38; overall prevalence: n = 26 [14.9%], n = 148 [85.1%]). Tumour location and histology are annotated.
Extended Data Fig. 3 PFS and OS with durvalumab-ceralasertib and with durvalumab plus olaparib, danvatirsen or oleclumab in patient subgroups defined by presence or absence of adverse prognostic factors.
Kaplan–Meier analyses of (left) PFS and (right) OS in patients with a, primary resistance, b, acquired resistance, c, PD-L1-negative tumours, d, PD-L1-positive tumours, e, squamous histology, f, non-squamous histology, and g, STK11 mutations.
Extended Data Fig. 4 PFS and OS with durvalumab-ceralasertib in patient subgroups defined by presence or absence of adverse prognostic factors.
Kaplan–Meier analyses of (left) PFS and (right) OS with durvalumab-ceralasertib in patients with a, primary vs acquired resistance, b, PD-L1-negative vs -positive status, and c, squamous cell carcinoma vs non-squamous histology, and in patients d, with or without bone/liver metastases, e, with high or low tumour mutational burden, and according to f, STK11 status and g, KRAS status (variant or wild-type).
Supplementary information
Supplementary Information
Supplementary Tables 1–13 and Fig. 1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Besse, B., Pons-Tostivint, E., Park, K. et al. Biomarker-directed targeted therapy plus durvalumab in advanced non-small-cell lung cancer: a phase 2 umbrella trial. Nat Med 30, 716–729 (2024). https://doi.org/10.1038/s41591-024-02808-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-024-02808-y
This article is cited by
-
The ATR inhibitor ceralasertib potentiates cancer checkpoint immunotherapy by regulating the tumor microenvironment
Nature Communications (2024)