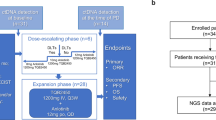
Abstract
INTRIGUE was an open-label, phase 3 study in adult patients with advanced gastrointestinal stromal tumor who had disease progression on or intolerance to imatinib and who were randomized to once-daily ripretinib 150 mg or sunitinib 50 mg. In the primary analysis, progression-free survival (PFS) with ripretinib was not superior to sunitinib. In clinical and nonclinical studies, ripretinib and sunitinib have demonstrated differential activity based on the exon location of KIT mutations. Therefore, we hypothesized that mutational analysis using circulating tumor DNA (ctDNA) might provide further insight. In this exploratory analysis (N = 362), baseline peripheral whole blood was analyzed by a 74-gene ctDNA next-generation sequencing–based assay. ctDNA was detected in 280/362 (77%) samples with KIT mutations in 213/362 patients (59%). Imatinib-resistant mutations were found in the KIT ATP-binding pocket (exons 13/14) and activation loop (exons 17/18). Mutational subgroup assessment showed 2 mutually exclusive populations with differential treatment effects. Patients with only KIT exon 11 + 13/14 mutations (ripretinib, n = 21; sunitinib, n = 20) had better PFS with sunitinib versus ripretinib (median, 15.0 versus 4.0 months). Patients with only KIT exon 11 + 17/18 mutations (ripretinib, n = 27; sunitinib, n = 25) had better PFS with ripretinib versus sunitinib (median, 14.2 versus 1.5 months). The results of this exploratory analysis suggest ctDNA sequencing may improve the prediction of the efficacy of single-drug therapies and support further evaluation of ripretinib in patients with KIT exon 11 + 17/18 mutations. ClinicalTrials.gov identifier: NCT03673501.
Similar content being viewed by others
Main
Gastrointestinal stromal tumor (GIST) is the most common gastrointestinal sarcoma, with approximately 80% of cases driven by mutations in KIT, and up to 10% by mutations in platelet-derived growth factor receptor α (PDGFRA)1,2,3. Imatinib, a KIT/PDGFRA tyrosine kinase inhibitor (TKI), is an effective first-line therapy for patients with advanced GIST; however, most patients ultimately develop disease progression due to secondary resistance mutations4,5,6,7,8,9,10. Approximately 90% of patients with KIT-mutant GIST who had disease progression on imatinib harbor newly acquired secondary KIT mutations, which most commonly appear in the ATP-binding pocket (encoded by exons 13/14) and/or activation loop (exons 17/18)11,12,13,14,15.
Sunitinib is the approved second-line therapy for patients with advanced GIST following progression on or intolerance to imatinib16. In the registrational phase 3 trial, patients treated with sunitinib demonstrated an overall median progression-free survival (PFS) of 5.6 months. However, there was no analysis of secondary mutations in the phase 3 trial, and sunitinib has demonstrated differential efficacy dependent on the location of imatinib-resistant KIT mutations11,17. In a phase 1/2 study (NCT00457743), the median PFS for sunitinib was 7.8 months in patients harboring secondary resistance mutations in the KIT ATP-binding pocket compared with 2.3 months in patients who had mutations in the activation loop11.
Ripretinib, a switch-control TKI, is approved for adult patients with advanced GIST who have received prior treatment with three or more TKIs, including imatinib, based on the results of the phase 3 INVICTUS study18,19. When compared with sunitinib in the phase 3 INTRIGUE trial, ripretinib demonstrated similar efficacy in patients who had disease progression on or were intolerant to imatinib in the KIT exon 11 intent-to-treat (ITT; median PFS, 8.3 versus 7.0 months, respectively; P = 0.36) and overall ITT populations (median PFS, 8.0 versus 8.3 months, respectively; nominal P = 0.72), suggesting that ripretinib demonstrated comparable efficacy to sunitinib as a second-line therapy20. Ripretinib also demonstrated a more favorable safety profile compared with sunitinib, with fewer patients experiencing grade 3/4 treatment-emergent adverse events (TEAEs)20. Based on these primary results from the INTRIGUE trial, ripretinib was recently included in the National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology for GIST (version 1.2023) as a preferred second-line regimen for patients with advanced GIST who are intolerant to sunitinib20,21. As fourth-line or later therapy, PFS with ripretinib was longer than placebo in all assessed mutational subgroups (KIT exons 9, 11, 13 and 17), suggesting broad activity in this later-line setting, irrespective of baseline mutation status22.
Tumor tissue obtained through biopsy has been the predominant source for mutational analysis in cancer; however, circulating tumor DNA (ctDNA) analysis is becoming more common23,24. Despite the limitations of using ctDNA (e.g., sample processing issues, assay specificity, low shedding disease), these analyses may provide more comprehensive information reflective of systemic tumor burden rather than the limited areas sampled by tissue biopsy24,25,26. Given the differential activity of TKIs depending on the location of KIT mutations as well as the poor activity of sunitinib in patients with secondary KIT exon 17/18 mutations, we hypothesized that further investigation by mutational subgroup using ctDNA could provide more insight into the efficacy of these agents as second-line therapies. In this prespecified exploratory analysis from INTRIGUE, we present the landscape of KIT mutations at the onset of imatinib failure and evaluate the efficacy of ripretinib versus sunitinib in patients with advanced GIST according to baseline KIT mutation status as determined by ctDNA analysis.
Results
ctDNA sample evaluability and landscape of KIT mutations
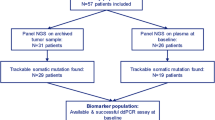
Of 453 patients in the overall ITT population, ctDNA was analyzed for 362 (80%; Fig. 1). A total of 280/362 patients (77%) had detectable ctDNA for single-nucleotide variants (SNVs) and/or insertions/deletions (INDELs); 213/362 (59%) had detectable KIT mutations (Fig. 1). The observed KIT mutations included KIT exon 9 (n = 36/213 (16.9%)), exon 11 (n = 157/213 (73.7%)), exons 13/14 (n = 81/213 (38.0%)) and exons 17/18 (n = 89/213 (41.8%)), with patients belonging to more than one group if they harbored multiple mutations (Fig. 1). Most patients harbored one or two KIT mutations (162 (76%)), with 6 patients exhibiting seven or more KIT mutations (Extended Data Fig. 1). Primary KIT exon 9 mutations were detected in 36 patients, with the most common being the AY duplication at codons 502–503 (n = 33 mutations); 157 patients had KIT exon 11 primary mutations, with codons 557–558 being the most impacted (n = 82 mutations; Extended Data Fig. 2a).
Patients are included in multiple groups if they had more than one mutation; patients can have multiple mutations in the same exon. Groups under each of the categories (KIT exon 9, 11, 13/14 or 17/18) are not mutually exclusive, and patients may appear in more than one box. Bold indicates patients who were included in the analysis populations for the current manuscript. CNV, copy number variation; QC, quality control; R, ripretinib; S, sunitinib. actDNA detected only for SNV/INDEL; two patients had CNV-only mutations and were categorized as ctDNA not detected.
Most patients had secondary resistance mutations in KIT exon 13 and/or exon 17 (Extended Data Fig. 2b). Overall, 42 unique secondary resistance mutations were observed in the KIT ATP-binding pocket (exons 13/14) and activation loop (exons 17/18; Fig. 2). The most common secondary resistance mutation was the V654A substitution in exon 13 (n = 65) followed by the Y823D (n = 37) and N822K (n = 26) substitutions in exon 17 (Fig. 2).
This plot illustrates the number of mutations; each patient could have multiple mutations. The letters in the bubbles and in front of each listed codon represent amino acids. A, alanine; ATP, adenosine triphosphate; C, cysteine; D, aspartic acid; E, glutamic acid; F, phenylalanine; G, glycine; H, histidine; I, isoleucine; K, lysine; L, leucine; M, methionine; N, asparagine; P, proline; R, arginine; S, serine; T, threonine; V, valine; Y, tyrosine. aE640_L641delinsD. bRipretinib: R815_D816delinsN; sunitinib: R815_D816delinsK.
When looking at median PFS by mutation subgroup, two diametrically opposed populations were evident (Fig. 3). Differential treatment effects were observed in patients with primary KIT exon 11 mutations with imatinib-resistant mutations exclusively in exons 13/14 (41/362 (11%)) and in patients with primary KIT exon 11 mutations with imatinib-resistant mutations exclusively in exons 17/18 (52/362 (14%)). Based on these findings, outcome results for these two mutually exclusive, diametrically opposed populations will be presented in the current article.
Data are represented as hazard ratio (HR) ± 95% confidence interval (CI). PFS was summarized using the Kaplan-Meier method with associated two-sided 95% CIs calculated using the Brookmeyer and Crowley method. HRs were obtained from the unstratified Cox proportional hazard model. Nine patients were included in multiple groups, including one patient with mutations in KIT exon 9 and KIT exon 11 and eight patients with mutations in KIT exon 11 and PDGFRA; the exon 11 + 13/14-only group excludes patients with mutations in KIT exons 9, 17 and 18; the exon 11 + 17/18-only group excludes patients with mutations in KIT exons 9, 13 and 14. Data cutoff: 1 September 2021.
Patients
Demographics and clinical characteristics for the overall ITT population were published previously20. Baseline demographic and clinical characteristics were well balanced between the KIT exon 11 + 13/14 and KIT exon 11 + 17/18 populations and between treatment arms (Table 1). The median age was 59.0 and 60.0 years in the KIT exon 11 + 13/14 and KIT exon 11 + 17/18 populations, respectively. Race was self-reported, and most patients were White males from North America or Europe (Table 1).
Efficacy
In the KIT exon 11 + 13/14 population, sunitinib demonstrated improved PFS compared with ripretinib (median, 15.0 versus 4.0 months; HR, 3.94; 95% CI, 1.71–9.11; nominal P = 0.0005; Figs. 3 and 4a). Conversely, ripretinib demonstrated improved PFS compared with sunitinib in patients with KIT exon 11 + 17/18 mutations (median, 14.2 versus 1.5 months; HR, 0.22; 95% CI, 0.11–0.44; nominal P < 0.0001; Figs. 3 and 4b). These results remained robust when accounting for multiple treatment comparisons across mutational subgroups, with a significant interaction between treatment and mutational subgroup (nominal P < 0.0001; Extended Data Table 1).
PFS was summarized using the Kaplan-Meier method with associated two-sided 95% CIs calculated using the Brookmeyer and Crowley method. HRs and P values were obtained from the unstratified Cox proportional hazard model and two-sided unstratified log-rank tests, respectively. Data cutoff: 1 September 2021. P values are nominal. NE, not estimable.
Similar to the PFS for the overall ITT and KIT exon 11 ITT populations in the primary analysis, PFS rates for the total (all samples analyzed) and any KIT exon 11 groups were similar between treatment arms (Fig. 3)20. PFS was better with sunitinib versus ripretinib for patients with only baseline KIT exon 11 mutations (median, 16.3 versus 2.2 months; HR, 2.24; 95% CI, 0.99–5.09; nominal P = 0.0460; Fig. 3). In patients with co-occurring imatinib-resistant secondary mutations in both the KIT ATP-binding pocket and activation loop (n = 22/362 (6%)), no difference was revealed between sunitinib and ripretinib (HR, 1.07; 95% CI, 0.41–2.84; nominal P = 0.8843; Fig. 3).
In the KIT exon 11 + 13/14 population, the objective response rate (ORR) was 9.5% with ripretinib versus 15.0% with sunitinib (response difference (RD), −5.5%; 95% CI, −27.6 to 16.2; nominal P = 0.5922; Fig. 5a,b). A higher ORR was observed with ripretinib versus sunitinib in the KIT exon 11 + 17/18 population (44.4% versus 0%; RD, 44.4%; 95% CI, 23.0–62.7; nominal P = 0.0001; Fig. 5c,d). Across both treatment arms, overall survival (OS) event rates were 51.2% and 50.0% in the KIT exon 11 + 13/14 and KIT exon 11 + 17/18 populations, respectively. Median OS was not reached for patients receiving sunitinib in the KIT exon 11 + 13/14 population, whereas the median OS for patients receiving ripretinib was 24.5 months (HR, 1.75; 95% CI, 0.72–4.24; nominal P = 0.2085; Extended Data Fig. 3a) with a median follow-up of 24.1 and 30.7 months for sunitinib and ripretinib, respectively. Improved OS was observed with ripretinib versus sunitinib in the KIT exon 11 + 17/18 population (median, not reached versus 17.5 months; HR, 0.34; 95% CI, 0.15 to 0.76; nominal P = 0.0061; Extended Data Fig. 3b) with a median follow-up of 29.7 months for ripretinib and 31.4 months for sunitinib. These results remained robust when accounting for multiple treatment comparisons across mutational subgroups, with a significant interaction between treatment and mutational subgroup (nominal P = 0.0179; Extended Data Table 2).
a,b, Patients with KIT exon 11 + 13/14 mutations. c,d, Patients with KIT exon 11 + 17/18 mutations. Data cutoff: 1 September 2021. Dotted line at 20% represents the threshold for PD; dotted line at −30% represents threshold for PR. DOR, duration of response; NE, not evaluable; PD, progressive disease; PR, partial response; SD, stable disease.
Safety and follow-up therapies
Median treatment duration for ripretinib versus sunitinib in the KIT exon 11 + 13/14 population was 4.6 versus 9.5 months, respectively. In the KIT exon 11 + 17/18 population, median treatment duration was 14.0 versus 3.0 months for patients receiving ripretinib versus sunitinib, respectively (Extended Data Table 3). The observed safety profile appeared to be consistent with the primary analysis. There were more grade 3/4 drug-related TEAEs in patients receiving sunitinib versus ripretinib (KIT exon 11 + 13/14: 50.0% versus 28.6%; KIT exon 11 + 17/18: 50.0% versus 33.3%, respectively; Extended Data Table 3). In the KIT exon 11 + 13/14 population, more patients receiving sunitinib versus ripretinib underwent dose interruptions and reductions due to any TEAE (30.0% versus 23.8% and 45.0% versus 4.8%, respectively; Extended Data Table 3). Conversely, in the KIT exon 11 + 17/18 population, more patients receiving ripretinib versus sunitinib underwent dose interruptions and dose reductions due to any TEAE (59.3% versus 41.7% and 37.0% versus 29.2%, respectively). However, when looking at the number of dose interruptions and reductions due to any TEAE in the first 12 weeks of the study, the proportions were either higher with sunitinib compared with ripretinib or comparable between the 2 arms (Extended Data Table 3). The most common TEAE of any grade observed with ripretinib was alopecia, regardless of mutational subgroup (KIT exon 11 + 13/14: 66.7%; KIT exon 11 + 17/18: 77.8%); the most common TEAEs with sunitinib in the KIT exon 11 + 13/14 and 11 + 17/18 populations were palmar-plantar erythrodysesthesia syndrome (60.0%) and hypertension (50.0%), respectively (Extended Data Table 4). Anticancer therapies received following discontinuation of study treatment can be found in Extended Data Table 5.
Discussion
This exploratory analysis from the phase 3 INTRIGUE trial in pretreated, advanced GIST demonstrates the potential value of ctDNA next-generation sequencing (NGS)-based analysis of imatinib-resistant secondary KIT mutations to select second-line treatment. In this analysis, there were 42 unique mutations in the KIT ATP-binding pocket (exons 13/14) and activation loop (exons 17/18). The vast majority of these variants are known to cause imatinib resistance, but some of the novel variants with uncertain significance may not. In patients with the most common class of primary driver mutation in GIST (KIT exon 11 mutation), imatinib-resistant secondary mutations in the KIT ATP-binding pocket correlated with clinical benefit from sunitinib versus ripretinib (median PFS, 15.0 versus 4.0 months, respectively; P = 0.0005), whereas secondary mutations in the KIT activation loop indicated clinical benefit from ripretinib but not sunitinib (median PFS, 14.2 versus 1.5 months, respectively; P < 0.0001). Although these results are limited by the exploratory nature of the analysis, the differences in these populations were robust when accounting for multiple treatment comparisons across mutational subgroups with or without adjustment for different baseline characteristics.
Although mutational testing is strongly recommended for optimal therapy of patients with treatment-naïve, advanced GIST before initiating therapy with TKIs21, it is only performed in a minority of patients in the United States27. The primary genotype determines selection of drug (and dose) for imatinib (KIT exon 11 versus KIT exon 9), as well as avapritinib (PDGFRA exon 18 D842V mutation)1,28,29 and NTRK and BRAF inhibitors for patients with activating mutations in these kinases30,31,32. However, other than baseline mutation testing, there are limited studies supporting routine analysis of secondary mutations to optimize the treatment decision for the next line of therapy15. At most GIST centers, patients with KIT-mutant GIST are treated sequentially with imatinib, sunitinib, regorafenib and ripretinib, as first- to fourth-line therapies, based on progression or intolerance during the previous line of the therapy21. In the primary results from the INTRIGUE study, ripretinib was not superior to sunitinib as a second-line therapy in terms of PFS in a molecularly unselected population20. The current exploratory analysis, however, suggests that ctDNA could identify a molecular subset of patients who may preferentially benefit from second-line treatment with ripretinib rather than with the recommended second-line therapy, sunitinib21.
To date, mutational analysis has been predominantly performed on tissue biopsy samples; however, tissue biopsies are an invasive procedure that sample a portion of a single-tumor lesion, and multiple biopsies within and/or across lesions are not justifiable in routine clinical practice23. Plasma ctDNA analysis can theoretically overcome these limitations, with easy access to blood and the potential to reflect the full mutational burden across multiple metastatic sites and identify patients who might benefit from specific cancer therapies25,26. Some reports indicate low ctDNA shedding in GIST; however, higher rates of ctDNA detection were observed in active, metastatic disease15,33,34, and the current study demonstrated a high rate of ctDNA detection in patients with advanced GIST previously treated with imatinib (280/362; 77%). A previous study demonstrated good concordance between ctDNA and NGS testing from tumor tissue in a small cohort of patients with metastatic GIST15. Furthermore, imatinib-resistant mutations have been detected in ctDNA samples that were not observed in tissue biopsies, suggesting that ctDNA assays may allow physicians and researchers to effectively monitor secondary resistance mutations and treatment in advanced GIST22,35,36. To this end, these data may support a ctDNA-guided treatment approach in GIST using a sensitive and minimally invasive test and require further investigation in prospective trials.
The differential activity of sunitinib against imatinib-resistant mutations in the KIT ATP-binding pocket and activation loop was previously documented in both clinical and nonclinical studies11,37,38. In a nonrandomized, single-arm trial evaluating sunitinib in patients with advanced GIST, imatinib-resistant secondary mutations within the KIT activation loop (detected in single-tumor biopsies) were associated with rapid clinical progression (median PFS, 2.3 months), whereas PFS was significantly longer for patients with secondary mutations in the KIT ATP-binding pocket (median PFS, 7.8 months; P = 0.0157)11. We hypothesized that ctDNA could be particularly helpful in determining effective single-drug treatment approaches as it appears there is a subset of patients with advanced GIST who may not benefit from second-line treatment with sunitinib.
Preclinically, ripretinib inhibited a broad panel of KIT mutants in GIST and non-GIST cell lines, including many of the common primary and secondary resistance mutations observed in patients with advanced GIST38. Ripretinib was less effective, however, against secondary mutations in the KIT ATP-binding pocket than in the activation loop, regardless of primary mutation (KIT exon 11 or 9). In contrast, ripretinib demonstrated clinical activity compared with placebo independent of baseline mutation status in patients with fourth-line advanced GIST, including in a subgroup harboring KIT ATP-binding pocket mutations (KIT exon 13)22. However, this subgroup included any patient with a KIT exon 13 mutation regardless of additional activation loop mutations, and the study did not have an active comparator arm. In the current study, patients harboring co-occurring mutations in the ATP-binding pocket and activation loop (KIT exons 11 + 13/14 + 17/18) performed similarly irrespective of treatment assignment. However, further investigation is warranted due to the small numbers of patients (11 patients in each treatment arm).
Based on the current findings, a phase 3, randomized, multicenter, open-label study evaluating ripretinib versus sunitinib in patients with advanced GIST previously treated with imatinib who harbor KIT exon 11 + 17 and/or 18 mutations (without co-occurring mutations in KIT exons 9, 13 or 14) is ongoing (INSIGHT; NCT05734105). In this follow-up phase 3 study, ripretinib was granted breakthrough therapy designation by the US Food and Drug Administration. INSIGHT aims to confirm not only the PFS observed with ripretinib in patients with KIT exon 11 + 17/18 mutations (activation loop) from this exploratory analysis, but also the response rate (ORR, 44.4%), which was almost three times the ORR observed with sunitinib in patients with KIT exon 11 + 13/14 mutations (ATP-binding pocket; ORR, 15.0%). This finding could be explained by the idea that sunitinib was primarily developed as a potent inhibitor of vascular endothelial growth factor receptor, whereas ripretinib was optimized to inhibit activated KIT (as opposed to competing with ATP binding to the kinase) to decrease unwanted toxicity38,39. This difference in response could reflect varied levels of kinase inhibition by ripretinib versus sunitinib against drug-sensitive mutations; however, no on-treatment biopsies were performed to confirm this hypothesis.
Limitations of the current study include the exploratory nature of the analysis; as such, all P values reported are nominal and no statistical significance can be claimed or cited in clinical practice. Additionally, there were low patient numbers in some mutational subgroups, making it difficult to interpret some outcomes. There also exists a subset of patients who do not have detectable ctDNA, which does not allow for the personalized treatment approaches proposed in this report. In addition, challenges associated with the technology, such as variables influencing ctDNA stability and sample processing, could contribute to decreased assay sensitivity24. Finally, the current study only evaluated imatinib-resistant secondary KIT mutations, and further investigation would be necessary to identify any KIT-independent resistance mechanisms.
In conclusion, in this cohort, patients whose ctDNA contained primary KIT exon 11 mutations plus secondary mutations restricted to KIT exons 17/18 demonstrated greater benefit from ripretinib versus sunitinib, with all the limitations of an exploratory biomarker analysis. In contrast, patients with the same primary mutation (KIT exon 11) and secondary mutations restricted to KIT exons 13/14 demonstrated greater benefit from sunitinib versus ripretinib. First and foremost, our data suggest that ctDNA analysis may represent a powerful, non-invasive diagnostic tool to identify subgroups of patients with advanced GIST who experienced disease progression on imatinib that may have prolonged clinical benefit from a single TKI therapeutic approach. To this end, ctDNA analysis may broadly determine the heterogeneity of resistance for an individual patient compared with a tissue biopsy, which provides information on only a single lesion. Further investigation of the efficacy of ripretinib as a second-line treatment is required and ongoing in the phase 3 INSIGHT trial (NCT05734105).
Methods
Study design
The INTRIGUE trial was conducted in accordance with the Declaration of Helsinki and International Council for Harmonisation Guidelines for Good Clinical Practice. The protocol, protocol amendments and informed consent documents were approved by a central institutional review board (WCG IRB, Puyallup, WA), as well as the institutional review board or ethics committee at each site (Supplementary Information) and by appropriate regulatory authorities. A list of all investigational sites for the INTRIGUE trial was published previously20. All patients provided written informed consent at enrollment. Participants were not compensated for participation.
INTRIGUE (NCT03673501) is a randomized, open-label, global, multicenter, phase 3 study comparing efficacy and safety of ripretinib versus sunitinib in patients with advanced GIST who had disease progression on or were intolerant to first-line treatment with imatinib. Patients were stratified by mutational status via tissue biopsy pathology report (KIT exon 11, KIT exon 9, KIT/PDGFRA wild-type and other KIT mutations (other than exons 9 or 11)/PDGFRA mutations) and by imatinib intolerance. Patient sex was self-reported and was not considered in the study design. Patients were randomized (1:1) to receive once-daily ripretinib 150 mg (continuous dosing) or once-daily sunitinib 50 mg (4 weeks on/2 weeks off in 6-week cycles). Crossover was not allowed. The study design and patient disposition were published previously20.
Eligibility criteria
Eligible patients were ≥18 years and had histologically confirmed GIST with one or more measurable lesions by modified Response Evaluation Criteria in Solid Tumors version 1.1 (mRECIST v1.1)40 criteria within 21 days before receiving study drug. Eligible patients provided an archival tissue sample and pathology report detailing KIT/PDGFRA mutation status by tissue-based PCR or any DNA sequencing analysis, had disease progression with imatinib or demonstrated imatinib intolerance, discontinued imatinib treatment 10 days before the first dose of study drug and had an Eastern Cooperative Oncology Group Performance Status ≤2 with acceptable organ function and bone marrow reserve. Full inclusion/exclusion criteria were published previously20.
ctDNA analysis
In this prespecified exploratory analysis, baseline (cycle 1 day 1) peripheral whole blood was collected in 10-mL Streck cell-free DNA blood collection tubes and shipped to central laboratories for plasma isolation. DNA extraction was performed by Guardant Health and samples were analyzed using Guardant360 (a 74-gene ctDNA NGS–based assay). This assay has a reported 99.6% specificity and 85.0% sensitivity when compared with tissue-based NGS41. SNVs and small INDELs can be reported as low as 0.04% and 0.02%, respectively42. Mean and median mutant allele frequencies for driver genes KIT and PDGFRA can be found in Extended Data Table 6.
Patients in the KIT exon 11 + 13/14 population have primary mutations in KIT exon 11 and secondary resistance mutations only in the KIT ATP-binding pocket (excludes patients with mutations in KIT exons 9, 17 and 18). Patients in the KIT exon 11 + 17/18 population have primary mutations in KIT exon 11 and secondary resistance mutations only in the KIT activation loop (excludes patients with mutations in KIT exons 9, 13 and 14). The current exploratory analysis does not include mutational information by tumor biopsy and only provides detailed outcome data for patients with typical secondary KIT mutations (ATP-binding pocket and activation loop).
Outcomes
The primary efficacy endpoint for the INTRIGUE trial was PFS by independent radiologic review (IRR) using mRECIST v1.1; key secondary endpoints were ORR by IRR using mRECIST v1.1 and OS20. In the current prespecified exploratory analysis, the baseline mutational landscape was characterized by analyzing the frequency of imatinib-resistant secondary KIT mutations in the ATP-binding pocket (exons 13/14) and activation loop (exons 17/18) by position and codon and determining the number of mutations in each patient. Mutational biomarkers that may correlate with treatment response were assessed via PFS, ORR and OS, which were the prespecified, protocol-defined endpoints for the INTRIGUE trial. Safety data are also reported. Data cutoff was 1 September 2021 for all outcomes except OS, which had an updated data cutoff of 1 September 2022.
Statistical analyses
Statistics and reproducibility
In the primary study, patients were randomized (1:1) to receive once-daily ripretinib or once-daily sunitinib. The investigators were not blinded to allocation during experiments and outcome assessment. The endpoints of PFS and ORR were based on IRR and the independent reviewer was blinded to treatment assignment. For this exploratory ctDNA analysis, no statistical method was used to predetermine sample size. The ctDNA assay used all the baseline sample and cannot be reproduced. Of the 374 patient samples received, 12 failed initial quality control review and were not analyzed. No data from the 362 analyzed samples were excluded.
Statistical tests
Time-to-event data were summarized using the Kaplan-Meier method with associated two-sided 95% CIs calculated using the Brookmeyer and Crowley method. HRs and P values were obtained from the unstratified Cox proportional hazard model and two-sided unstratified log-rank tests, respectively. ORR was analyzed by the chi-square test for association between treatment and ORR; the 95% CI of the RD was calculated using the unstratified Newcombe method. Descriptive statistics were used to summarize safety data. All P values reported for this exploratory analysis are nominal. Statistical analyses were done with SAS (version 9.4).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The redacted study protocol for the INTRIGUE trial was previously published and can be accessed here: https://ascopubs.org/doi/suppl/10.1200/JCO.22.00294/suppl_file/protocol_JCO.22.00294.pdf. The ctDNA dataset contains person-sensitive data and is not broadly available due to privacy laws. Qualified scientific and medical researchers can make requests for individual participant data that underlie the results reported in this article, after de-identification, at info@deciphera.com. Proposals for data will be evaluated and approved by Deciphera in its sole discretion. All approved researchers must sign a data access agreement before accessing the data. Data will be available as soon as possible but no later than within 1 year of the acceptance of the article for publication and for 3 years after article publication. Deciphera will not share data from identified participants or a data dictionary.
References
Corless, C. L. et al. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J. Clin. Oncol. 23, 5357–5364 (2005).
Rubin, B. P., Heinrich, M. C. & Corless, C. L. Gastrointestinal stromal tumour. Lancet 369, 1731–1741 (2007).
Vallilas, C. et al. Gastrointestinal stromal tumors (GISTs): novel therapeutic strategies with immunotherapy and small molecules. Int. J. Mol. Sci. 22, 493 (2021).
Antonescu, C. R. et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin. Cancer Res. 11, 4182–4190 (2005).
Wardelmann, E. et al. Acquired resistance to imatinib in gastrointestinal stromal tumours caused by multiple KIT mutations. Lancet Oncol. 6, 249–251 (2005).
Wardelmann, E. et al. Polyclonal evolution of multiple secondary KIT mutations in gastrointestinal stromal tumors under treatment with imatinib mesylate. Clin. Cancer Res. 12, 1743–1749 (2006).
Blanke, C. D. et al. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the kit receptor tyrosine kinase: S0033. J. Clin. Oncol. 26, 626–632 (2008).
Schaefer, I. M., DeMatteo, R. P. & Serrano, C. The GIST of advances in treatment of advanced gastrointestinal stromal tumor. Am. Soc. Clin. Oncol. Educ. Book 42, 1–15 (2022).
Novartis Pharmaceuticals Corporation. Gleevec (imatinib mesylate) tablets, for oral use. Prescribing information. https://www.novartis.com/us-en/sites/novartis_us/files/gleevec_tabs.pdf (2022).
Chen, L. L. et al. A missense mutation in KIT kinase domain 1 correlates with imatinib resistance in gastrointestinal stromal tumors. Cancer Res. 64, 5913–5919 (2004).
Heinrich, M. C. et al. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J. Clin. Oncol. 26, 5352–5359 (2008).
Liegl, B. et al. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J. Pathol. 216, 64–74 (2008).
Serrano, C. & George, S. Gastrointestinal stromal tumor: challenges and opportunities for a new decade. Clin. Cancer Res. 26, 5078–5085 (2020).
Heinrich, M. C. et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J. Clin. Oncol. 24, 4764–4774 (2006).
Arshad, J. et al. Utility of circulating tumor DNA in the management of patients with GI stromal tumor: analysis of 243 patients. JCO Precis. Oncol. 4, 66–73 (2020).
Pfizer Labs. Sutent (sunitinib malate) capsules, for oral use. Prescribing information. https://labeling.pfizer.com/showlabeling.aspx?id=607 (2021).
Demetri, G. D. et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet 368, 1329–1338 (2006).
Blay, J. Y. et al. Ripretinib in patients with advanced gastrointestinal stromal tumours (INVICTUS): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 21, 923–934 (2020).
Deciphera Pharmaceuticals LLC. QINLOCK® (ripretinib) tablets, for oral use. Prescribing information. https://www.qinlockhcp.com/Content/files/qinlock-prescribing-information.pdf (2023).
Bauer, S. et al. Ripretinib versus sunitinib in patients with advanced gastrointestinal stromal tumor after treatment with imatinib (INTRIGUE): A randomized, open-label, phase III trial. J. Clin. Oncol. 40, 3918–3928 (2022).
National Comprehensive Cancer Network. NCCN guidelines gastroinestinal stromal tumors (GISTs) version 1.2023. https://www.nccn.org/professionals/physician_gls/pdf/gist.pdf (2023).
Bauer, S. et al. Clinical activity of ripretinib in patients with advanced gastrointestinal stromal tumor harboring heterogeneous KIT/PDGFRA mutations in the phase III INVICTUS study. Clin. Cancer Res. 27, 6333–6342 (2021).
Nannini, M., Astolfi, A., Urbini, M., Biasco, G. & Pantaleo, M. A. Liquid biopsy in gastrointestinal stromal tumors: a novel approach. J. Transl. Med. 12, 210 (2014).
Chen, M. & Zhao, H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum. Genomics 13, 34 (2019).
Elazezy, M. & Joosse, S. A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 16, 370–378 (2018).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 36, 1631–1641 (2018).
Florindez, J. & Trent, J. Low frequency of mutation testing in the United States: an analysis of 3866 GIST patients. Am. J. Clin. Oncol. 43, 270–278 (2020).
Blueprint Medicines Corporation. AYVAKIT (avapritinib) tablets, for oral use. Prescribing information. https://www.blueprintmedicines.com/wp-content/uploads/uspi/AYVAKIT.pdf (2023).
Yoo, C. et al. Efficacy of imatinib in patients with platelet-derived growth factor receptor alpha-mutated gastrointestinal stromal tumors. Cancer Res. Treat. 48, 546–552 (2016).
Bayer Healthcare Pharmaceuticals. Vitrakvi (larotrectinib) tablets or solution, for oral use. Prescribing information. https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/211710s000lbl.pdf (2018).
Novartis Pharmaceuticals Corporation. Mekinist (trametinib) tablets, for oral use. Prescribing information. https://www.novartis.com/us-en/sites/novartis_us/files/mekinist.pdf (2023).
Novartis Pharmaceuticals Corporation. Tafinlar (dabrafenib) tablets, for oral use. Prescribing information. https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/202806s022lbl.pdf (2022).
Serrano, C. et al. Clinical value of next generation sequencing of plasma cell-free DNA in gastrointestinal stromal tumors. BMC Cancer 20, 99 (2020).
Ko, T. K. et al. Circulating tumor DNA mutations in progressive gastrointestinal stromal tumors identify biomarkers of treatment resistance and uncover potential therapeutic strategies. Front. Oncol. 12, 840843 (2022).
Xu, H. et al. Clinical application of circulating tumor DNA in the genetic analysis of patients with advanced GIST. Mol. Cancer Ther. 17, 290–296 (2018).
Demetri, G. D. et al. Mutational analysis of plasma DNA from patients (pts) in the phase III GRID study of regorafenib (REG) versus placebo (PL) in tyrosine kinase inhibitor (TKI)-refractory GIST: Correlating genotype with clinical outcomes. J. Clin. Oncol. 31, 10503–10503 (2013).
Serrano, C. et al. Complementary activity of tyrosine kinase inhibitors against secondary kit mutations in imatinib-resistant gastrointestinal stromal tumours. Br. J. Cancer 120, 612–620 (2019).
Smith, B. D. et al. Ripretinib (DCC-2618) is a switch control kinase inhibitor of a broad spectrum of oncogenic and drug-resistant KIT and PDGFRA variants. Cancer Cell 35, 738–751 e739 (2019).
Mendel, D. B. et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin. Cancer Res. 9, 327–337 (2003).
Demetri, G. D. et al. Efficacy and safety of regorafenib for advanced gastrointestinal stromal tumours after failure of imatinib and sunitinib (GRID): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 381, 295–302 (2013).
Lanman, R. B. et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 10, e0140712 (2015).
Odegaard, J. I. et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin. Cancer Res. 24, 3539–3549 (2018).
Acknowledgements
The INTRIGUE study was funded by Deciphera Pharmaceuticals, and as the sponsor, Deciphera contributed to the conception, design and analysis of the study. We thank the patients and their families and caregivers, the investigators and the investigational site staff of the INTRIGUE study. We also thank M. Kusi for her contributions to this manuscript and Guardant Health for processing plasma samples and providing the relevant methods. M.C.H. received partial salary support from the following sources: a research grant from the Jonathan David Foundation, a VA Merit Review Grant (I01BX005358) and NCI R21 grant (R21CA263400). Medical writing support was provided by L. Hanlon and M. Chakhtoura of AlphaBioCom, a Red Nucleus company, and was funded by Deciphera Pharmaceuticals, in compliance with international good publication practice guidelines.
Author information
Authors and Affiliations
Contributions
All authors had access to the data, participated in drafting and reviewing the manuscript and approved the final version for submission. M.C.H., R.L.J., S.G., H.G., P.S., M.v.M., J.R.Z., J.T., S.A., K.S., W.R., M.L.S., R.R.-S., J.Y.B. and S.B. participated in the conception and design of the study, along with the sponsor. M.C.H., R.L.J., S.G., H.G., P.S., M.v.M., J.R.Z., Y.-K.K., A.A.R., J.T., S.A., A.L.C., B.L.S., D.G., K.B., C.S., N.S., P.R., M.D., C.S., N.S., P.C., J.-Y.B. and S.B., along with the sponsor, participated in the collection of data. W.R. conducted the statistical analyses. Authors employed by Deciphera Pharmaceuticals contributed to the design and conception of the study, analyzed and interpreted the data in collaboration with all authors, participated in drafting and reviewing the manuscript and approved the final version for submission. All authors are accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
M.C.H. reports honoraria from Novartis; consulting/advisory roles for Blueprint Medicines, Deciphera, Novartis and Theseus Pharmaceuticals; patents/royalties/other intellectual property licensed to Novartis (institutional (Inst); treatment of GIST); and partial salary from a research grant from the Jonathan David Foundation, a VA Merit Review Grant (I01BX005358) and NCI R21 grant (R21CA263400). R.L.J. reports consulting/advisory roles for Lilly, Immune Design, Merck Serono, Adaptimmune, Daiichi Sankyo, Eisai, Morphotek, TRACON Pharma, Immodulon Therapeutics, Deciphera Pharmaceuticals, PharmaMar, Blueprint Medicines, Clinigen Group, Epizyme, Boehringer Ingelheim, Bayer, Karma Oncology, UpToDate, Athenex, Immunicum, Mundipharma, SpringWorks Therapeutics and SynOX; funding for travel/accommodations/expenses from PharmaMar; and research funding from GlaxoSmithKline (Inst). S.G. reports stock/other ownership interests from Abbott Laboratories; honoraria from CStone Pharmaceuticals; consulting or advisory roles for Blueprint Medicines, Deciphera, Bayer, Lilly, UpToDate, Research to Practice, MORE Health, Daiichi, KayoThera and Immunicum; research funding from Blueprint Medicines (Inst), Deciphera (Inst), Daiichi Sankyo RD Novare (Inst), Merck (Inst), Eisai (Inst), SpringWorks Therapeutics (Inst), TRACON Pharma (Inst), Theseus Pharmaceuticals (Inst) and BioAtla; patents/royalties/other intellectual property from UpToDate; expert testimony for Bayer; and other relationships with Research to Practice and WCG. H.G. reports research funding from Novartis (Inst), Ipsen (Inst), Deciphera (Inst) and Daiichi Sankyo (Inst). P.S. reports honoraria from Blueprint Medicines; consulting/advisory roles for Deciphera, Ellipses Pharma, Blueprint Medicines, Transgene, Exelixis, Boehringer Ingelheim, Studiecentrum voor Kernenergie (SCK CEN), SQZ Biotechnology, Adcendo, PharmaMar, Merck and Medpace; and research funding from CoBioRes NV (Inst), Eisai (Inst), G1 Therapeutics (Inst), PharmaMar (Inst), Genmab (Inst), Merck (Inst), Sartar Therapeutics (Inst) and ONA Therapeutics (Inst). M.v.M. reports consulting/advisory roles for Deciphera and GlaxoSmithKline; research funding from Novartis (Inst), Deciphera (Inst), Gradalis (Inst), Genmab (Inst), ASCO (Inst) and Solaris Health (Inst); patents/royalties/other intellectual property from Cell Line; and a relationship with the National Comprehensive Cancer Network. J.R.Z. reports a leadership role with ICON Group; stock/other ownership interests in Biomarin, Opthea, Amarin Corporation, Concert Pharmaceuticals, Frequency Therapeutics, Gilead Sciences, Madrigal Pharmaceuticals, UniQure, Zogenix, Orphazyme, Moderna Therapeutics, Twist Bioscience and Novavax; honoraria from Specialised Therapeutics, Merck Serono, Targovax, Halozyme, Gilead Sciences and Deciphera; consulting/advisory roles with Merck Serono, Targovax, Merck Sharp & Dohme, Halozyme, Lipotek, Specialised Therapeutics, Center for Emerging & Neglected Diseases (CEND), Deciphera, REVOLUTION MEDICINE, FivePHusion, Genor BioPharma, 1Globe Health Institute and Novotech; research funding from Merck Serono (Inst), Bristol Myers Squibb (Inst), AstraZeneca (Inst), Pfizer (Inst), IQVIA (Inst), Mylan (Inst), Ipsen (Inst), Eisai (Inst), Medtronic (Inst) and MSD Oncology (Inst); and funding for travel/accommodations/expenses from Merck Serono, AstraZeneca, Merck Sharp & Dohme, Deciphera and Sanofi. Y.-K.K. reports consulting/advisory roles with DAEHWA Pharmaceutical, Bristol Myers Squibb, Zymeworks, ALX Oncology, Amgen, Novartis, Macrogenics, Surface Oncology and Blueprint Medicines. A.A.R. reports consulting/advisory roles with Adaptimmune, Bayer, GlaxoSmithKline, Medison and Inhibrx; research funding from Deciphera, Karyopharm Therapeutics, Pfizer, Roche/Genentech, Bristol Myers Squibb, MedImmune, Amgen, GlaxoSmithKline, Blueprint Medicines, Merck, AbbVie, Adaptimmune, Iterion Therapeutics, Neoleukin Therapeutics, Daiichi Sankyo, Symphogen and Rain Therapeutics; and expert testimony for Medison. J.T. reports consulting/advisory roles for Blueprint Medicines, Deciphera, Daiichi Sankyo, Epizyme, Agios, C4 Therapeutics, Bayer, AADI Bioscience, LLC, Foghorn Therapeutics, Boehringer Ingelheim, Cogent Medicine and SERVIER. S.A. reports research funding from AB Science (Inst), TRACON Pharma (Inst), Bayer (Inst), Novartis (Inst), Lilly (Inst), Immune Design (Inst), Karyopharm Therapeutics (Inst), Epizyme (Inst), Blueprint Medicines (Inst), Genmab (Inst), CBA Pharma (Inst), Desmoid Tumor Research Foundation, Merck (Inst), Philogen (Inst), Gradalis (Inst), Deciphera (Inst), Takeda (Inst), Incyte (Inst), SpringWorks Therapeutics (Inst), Adaptimmune (Inst), Advenchen Laboratories (Inst), Bavarian Nordic (Inst), BTG (Inst), PTC Therapeutics (Inst), GlaxoSmithKline (Inst), FORMA Therapeutics (Inst), TRACON Pharma (Inst), Ayala Pharmaceuticals (Inst), Trillium Therapeutics (Inst), Boehringer Ingelheim (Inst), Salarius Pharmaceuticals (Inst), Theseus Pharmaceuticals (Inst), Monopar Therapeutics (Inst), C4 Therapeutics (Inst), InhibRx (Inst), Noxopharm (Inst) and Rain Therapeutics (Inst). A.L.C. reports honoraria from Bayer, PharmaMar and Deciphera. B.L.S. reports a consulting/advisory role for Deciphera (Inst); and research funding from Deciphera (Inst). D.G. reports honoraria from Sun Biopharma, Boehringer Ingelheim and AstraZeneca; consulting/advisory roles for Sun Biopharma, Seattle Genetics, AstraZeneca and Boehringer Ingelheim; and research funding from Amgen (Inst), Pfizer (Inst), Celgene (Inst), Bayer (Inst), Zucero Therapeutics (Inst) and Bristol Myers Squibb (Inst). K.B. reports honoraria from Novartis; and a consulting/advisory role for GlaxoSmithKline. C.S. reports research funding from Roche and Novartis; expert testimony for Pfizer; and funding for travel/accommodations/expenses from Novartis and Pfizer. N.S. reports consulting/advisory roles for Boehringer Ingelheim (Inst), Ellipses Pharma (Inst), Cogent Biosciences (Inst) and Luszana (Inst); and research funding from AstraZeneca/MedImmune (Inst), Bayer (Inst), Bristol Myers Squibb (Inst), Novartis (Inst), GlaxoSmithKline (Inst), Pfizer (Inst), Roche (Inst), Boehringer Ingelheim (Inst), Blueprint Medicines (Inst), Deciphera (Inst), Genentech (Inst), Merck Sharp & Dohme (Inst), Amgen (Inst), Merus (Inst), Incyte (Inst), AbbVie (Inst), Actuate Therapeutics (Inst), Sanofi (Inst), Cytovation (Inst), InteRNA (Inst), Array BioPharma (Inst), Cantargia AB (Inst), Taiho Pharmaceutical (Inst), Takeda (Inst), Ascendis Pharma (Inst), BridgeBio Pharma (Inst), CellCentric (Inst), Crescendo Biologics (Inst), Lilly (Inst), Exelixis (Inst), Janssen (Inst), Merck KGaA (Inst), Molecular Partners (Inst), Numab (Inst), Seattle Genetics (Inst), Cogent Biosciences (Inst), Kinnate Biopharma (Inst), Kling Biotherapeutics (Inst), Navire (Inst), Luszana (Inst), Relay Therapeutics (Inst), Revolution Medicines (Inst) and Dragonfly Therapeutics (Inst). P.R. reports honoraria from MSD, BMS and Pierre Fabre; advisory board roles for MSD, BMS, Pierre Fabre, Merck, Sanofi, Blueprint Medicines and Philogen; invited speaker roles for Merck, Sanofi and Novartis; research funding from Pfizer (Inst) and BMS (Inst); and nonfinancial interests as an invited speaker for the Polish Society of Surgical Oncology and the Polish Oncological Society (President Elect), and as an officer for ASCO. M.D. reports consulting/advisory roles for Adaptimmune, Deciphera and Epizyme; and speakers’ bureau participation for Deciphera. C.S. reports consulting/advisory roles for Deciphera, Blueprint Medicines, Immunicum and Cogent Biosciences; funding for travel/accommodations/expenses from PharmaMar, Pfizer, Bayer and Gilead Sciences; honoraria from Deciphera and PharmaMar; and research funding from Karyopharm Therapeutics (Inst) and IDRX (Inst). N.S. reports consulting/advisory roles for Deciphera, AADi, Blueprint Medicines, Bayer, Epizyme and Boehringer Ingelheim; stock/other ownership interests in Pfizer and Johnson & Johnson; and research funding from GlaxoSmithKline, Karyopharm Therapeutics, Deciphera, Ascentage Pharma, Daiichi Sankyo/Lilly and AstraZeneca/MedImmune (Inst). P.C. reports consulting/advisory roles for Deciphera, NewBay Pharma and Zai Lab; patents/royalties/other intellectual property from ORIC pharmaceuticals (immediate family member); stock/other ownership interests in ORIC pharmaceuticals (immediate family member); and research funding from Deciphera (Inst), Pfizer (Inst) and NewBay Pharma (Inst). W.R. reports employment with Deciphera; and stock and other ownership interests in Deciphera. K.S. reports employment with Deciphera (self) and Stablix (immediate family member); stock and other ownership interests in Deciphera (self) and Stablix (immediate family member); and a consulting/advisory role for Ipsen (immediate family member). H.A. reports employment with Deciphera; and stock and other ownership interests in Deciphera. M.L.S. reports employment with Deciphera; independent board of director position with Pieris Pharmaceuticals; leadership roles with Deciphera and Pieris Pharmaceuticals; stock/other ownership interests in Deciphera and Pieris Pharmaceuticals; and patents/royalties/other intellectual property from Acceleron Pharma. R.R.-S. reports employment with Deciphera; stock/other ownership interests in Deciphera and Immunogen; and patents/royalties/other intellectual property from Immunogen (inventor in three patents with Immunogen, transferred the rights to Immunogen, has not received (and will not receive) any royalties) and Deciphera (inventor in pending patents at Deciphera, transferred the rights to Deciphera, has not received (and will not receive) any royalties). J.-Y.B. reports a leadership role with Innate Pharma; honoraria from Roche, AstraZeneca, PharmaMar, MSD, BMS, Bayer, Ignyta, and Deciphera; consulting/advisory roles with Roche, PharmaMar, Blueprint Medicines, Bayer, Deciphera and Karyopharm Therapeutics; research funding from GlaxoSmithKline (Inst), PharmaMar (Inst), Novartis (Inst), Bayer (Inst), Roche (Inst), BMS (Inst), MSD (Inst), Deciphera (Inst), AstraZeneca (Inst) and OSE Pharma (Inst); and funding for travel/accommodations/expenses from Roche. S.B. reports honoraria from Novartis, Pfizer, Bayer, PharmaMar, GlaxoSmithKline and Deciphera; consulting/advisory roles with Blueprint Medicines, Bayer, Lilly, Deciphera, Nanobiotix, Daiichi Sankyo, Exelixis, Janssen-Cilag, ADC Therapeutics, Mundipharma, GlaxoSmithKline, Adcendo and Boehringer Ingelheim; research funding from Blueprint Medicines, Novartis and Incyte (Inst); and funding for travel/accommodations/expenses from PharmaMar.
Peer review
Peer review information
Nature Medicine thanks Amanda Blackford, Brian Van Tine, Dominic Rothwell, Toshirou Nishida, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Anna Maria Ranzoni, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Frequency of KIT mutations.
Includes both primary and secondary mutations. Only somatic KIT mutations considered; sunitinib arm includes 2 patients not treated. Total patients, N = 213. ctDNA, circulating tumor DNA.
Extended Data Fig. 2 Number of patients with KIT primary (A) and secondary resistance (B) mutations.
Data indicate number of patients with mutations at all affected codons; each patient can have multiple mutations. AA, amino acid.
Extended Data Fig. 3 Kaplan-Meier analysis of OS for patients treated with ripretinib or sunitinib in the KIT exon 11 + 13/14 (A) and KIT exon 11 + 17/18 (B) populations.
OS was summarized using the Kaplan-Meier method with associated 2-sided 95% CIs calculated using the Brookmeyer and Crowley method. HRs and P-values were obtained from the unstratified Cox proportional hazard model and 2-sided unstratified log-rank tests, respectively. Data cutoff: 1 September 2022. P-values are nominal. CI, confidence interval; HR, hazard ratio; NE, not estimable; OS, overall survival.
Supplementary information
Supplementary Information
Supplementary information.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Heinrich, M.C., Jones, R.L., George, S. et al. Ripretinib versus sunitinib in gastrointestinal stromal tumor: ctDNA biomarker analysis of the phase 3 INTRIGUE trial. Nat Med 30, 498–506 (2024). https://doi.org/10.1038/s41591-023-02734-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-023-02734-5