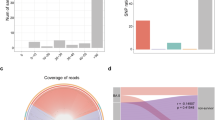
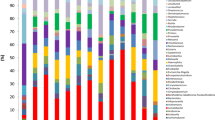
Abstract
Respiratory microbial dysbiosis is associated with acute respiratory distress syndrome (ARDS) and hospital-acquired pneumonia (HAP) in critically ill patients. However, we lack reproducible respiratory microbiome signatures that can increase our understanding of these conditions and potential treatments. Here, we analyze 16S rRNA sequencing data from 2,177 respiratory samples collected from 1,029 critically ill patients (21.7% with ARDS and 26.3% with HAP) and 327 healthy controls, sourced from 17 published studies. After data harmonization and pooling of individual patient data, we identified microbiota signatures associated with ARDS, HAP and prolonged mechanical ventilation. Microbiota signatures for HAP and prolonged mechanical ventilation were characterized by depletion of a core group of microbes typical of healthy respiratory samples, and the ARDS microbiota signature was distinguished by enrichment of potentially pathogenic respiratory microbes, including Pseudomonas and Staphylococcus. Using machine learning models, we identified clinically informative, three- and four-factor signatures that predicted ARDS, HAP and prolonged mechanical ventilation with relatively high accuracy (area under the curve of 0.751, 0.72 and 0.727, respectively). We validated the signatures in an independent prospective cohort of 136 patients on mechanical ventillation and found that patients with microbiome signatures associated with ARDS, HAP or prolonged mechanical ventilation had longer times to successful extubation than patients lacking these signatures (hazard ratios of 1.56 (95% confidence interval (CI) 1.07–2.27), 1.51 (95% CI 1.02–2.23) and 1.50 (95% CI 1.03–2.18), respectively). Thus, we defined and validated robust respiratory microbiome signatures associated with ARDS and HAP that may help to identify promising targets for microbiome therapeutic modulation in critically ill patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Correspondence and requests should be addressed to A.R. The 16S rRNA sequence dataset of the validation cohort has been deposited in a secure file by CHU Nantes (HAP2 data hub) and in the BioProject repository (PRJNA1020542). The data from the studies included in the meta-analysis can be found at figshare at https://figshare.com/s/46c402c6465c8a289c18, https://doi.org/10.6084/m9.figshare.3496412, https://doi.org/10.6084/m9.figshare.3496538, https://doi.org/10.6084/m9.figshare.3485201 and in the BioProject repository: PRJEB26875, PRJEB13056, PRJEB20913, PRJNA267584, SRP062137, PRJEB20665, PRJNA678854, PRJNA595346, SRP112361, PRJNA553560, PRJNA553560, SRP076183, PRJNA339755. The accession for each study is reported in Supplementary Tables 1–3.
Code availability
All analyses were performed using publicly available software and published code as described in the Methods. All custom codes generated for data analyses in this study are reported in the Methods.
References
He, Q. et al. The epidemiology and clinical outcomes of ventilator-associated events among 20,769 mechanically ventilated patients at intensive care units: an observational study. Crit. Care 25, 44 (2021).
Bellani, G. et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA 315, 788–800 (2016).
Roquilly, A. et al. Implementation of French recommendations for the prevention and the treatment of hospital-acquired pneumonia: a cluster-randomized trial. Clin. Infect. Dis. 73, e1601–e1610 (2020).
Magill, S. S. et al. Changes in prevalence of health care–associated infections in U.S. hospitals. N. Engl. J. Med. 379, 1732–1744 (2018).
Roquilly, A. et al. Interferon gamma-1b for the prevention of hospital-acquired pneumonia in critically ill patients: a phase 2, placebo-controlled randomized clinical trial. Intensive Care Med. 49, 530–544 (2023).
Luyt, C.-E. et al. Pulmonary infections complicating ARDS. Intensive Care Med. 46, 2168–2183 (2020).
Morris, A. & Flores, S. C. Study of the lung microbiome. Have we reached the end of the beginning?. Am. J. Resp. Crit. Care Med. 195, 15–16 (2017).
Charlson, E. S. et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Resp. Crit. Care Med. 184, 957–963 (2011).
Man, W. H., Piters, W. A. A., de, S. & Bogaert, D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 15, 259–270 (2017).
Dickson, R. P. et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann. Am. Thorac. Soc. 12, 821–830 (2015).
Dickson, R. P., Erb-Downward, J. R. & Huffnagle, G. B. Homeostasis and its disruption in the lung microbiome. Am. J. Physiol. Lung Cell. Mol. Physiol. 309, L1047–L1055 (2015).
Dickson, R. P. et al. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am. J. Resp. Crit. Care Med. 198, 497–508 (2018).
Dickson, R. P. et al. Lung microbiota predict clinical outcomes in critically ill patients. Am. J. Resp. Crit. Care Med. 201, 555–563 (2020).
Dickson, R. P. et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 1, nmicrobiol2016113 (2016).
Kelly, B. J. et al. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome 4, 7 (2016).
Kitsios, G. D. et al. Respiratory tract dysbiosis is associated with worse outcomes in mechanically ventilated patients. Am. J. Resp. Crit. Care Med. 202, 1666–1677 (2020).
Panzer, A. R. et al. Lung microbiota is related to smoking status and to development of acute respiratory distress syndrome in critically ill trauma patients. Am. J. Resp. Crit. Care Med. 197, 621–631 (2018).
Emonet, S. et al. Identification of respiratory microbiota markers in ventilator-associated pneumonia. Intensive Care Med. 45, 1082–1092 (2019).
Sommerstein, R. et al. Patterns in the longitudinal oropharyngeal microbiome evolution related to ventilator-associated pneumonia. Antimicrob. Resist. Infect. Control 8, 81 (2019).
Qi, X. et al. Lower respiratory tract microbial composition was diversified in Pseudomonas aeruginosa ventilator-associated pneumonia patients. Respir. Res. 19, 139 (2018).
Fromentin, M., Ricard, J.-D. & Roux, D. Respiratory microbiome in mechanically ventilated patients: a narrative review. Intensive Care Med. 47, 292–306 (2021).
Force, A. D. T. et al. Acute respiratory distress syndrome: the Berlin definition. JAMA 307, 2526–2533 (2012).
Torres, A. et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia. Eur. Respir. J. 50, 1700582 (2017).
Kalil, A. C. et al. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin. Infect. Dis. 63, e61–e111 (2016).
Dickson, R. P. et al. Bacterial topography of the healthy human lower respiratory tract. mBio 8, e02287–16 (2017).
Morris, A. et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Resp. Crit. Care Med. 187, 1067–1075 (2013).
Bassis, C. M. et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 6, e00037–15 (2015).
Chotirmall, S. H. et al. Therapeutic targeting of the respiratory microbiome. Am. J. Resp. Crit. Care Med. 206, 535–544 (2022).
François, B. et al. Efficacy and safety of suvratoxumab for prevention of Staphylococcus aureus ventilator-associated pneumonia (SAATELLITE): a multicentre, randomised, double-blind, placebo-controlled, parallel-group, phase 2 pilot trial. Lancet Infect. Dis. 21, 1313–1323 (2021).
Johnstone, J. et al. Effect of probiotics on incident ventilator-associated pneumonia in critically ill patients. JAMA 326, 1024–1033 (2021).
Litton, E. et al. Early and sustained Lactobacillus plantarum probiotic therapy in critical illness: the randomised, placebo-controlled, restoration of gut microflora in critical illness trial (ROCIT). Intensive Care Med. 47, 307–315 (2021).
Cheng, A. G. et al. Design, construction, and in vivo augmentation of a complex gut microbiome. Cell 185, e19 (2022).
Rangan, K. J. et al. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 353, 1434–1437 (2016).
Mitsi, E. et al. Nasal pneumococcal density is associated with microaspiration and heightened human alveolar macrophage responsiveness to bacterial pathogens. Am. J. Resp. Crit. Care Med. 201, 335–347 (2019).
Stewart, C. J. et al. Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis. A multiomic analysis. Am. J. Resp. Crit. Care Med. 196, 882–891 (2017).
Fatahi-Bafghi, M. Characterization of the Rothia spp. and their role in human clinical infections. Infect. Genet. Evol. 93, 104877 (2021).
Crémet, L. et al. Evaluation of the FilmArray® Pneumonia Plus panel for rapid diagnosis of hospital-acquired pneumonia in intensive care unit patients. Front. Microbiol. 11, 2080 (2020).
Pendleton, K. M. et al. Rapid pathogen identification in bacterial pneumonia using real-time metagenomics. Am. J. Resp. Crit. Care Med. 196, 1610–1612 (2017).
Guillotin, F. et al. Potential impact of rapid multiplex PCR on antimicrobial therapy guidance for ventilated hospital-acquired pneumonia in critically ill patients, a prospective observational clinical and economic study. Front. Cell. Infect. Microbiol. 12, 804611 (2022).
Moy, A.-C. et al. Performance evaluation of a PCR panel (FilmArray® Pneumonia Plus) for detection of respiratory bacterial pathogens in respiratory specimens: a systematic review and meta-analysis. Anaesth. Crit. Care Pain Med. 42, 101300 (2023).
Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689 (2019).
Lee, K. A. et al. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat. Med. 28, 535–544 (2022).
Chanderraj, R. et al. In critically ill patients, anti-anaerobic antibiotics increase risk of adverse clinical outcomes. Eur. Respir. J. 61, 2200910 (2023).
Kitsios, G. D. et al. The upper and lower respiratory tract microbiome in severe aspiration pneumonia. iScience 26, 106832 (2023).
Amir, A. et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191–16 (2017).
Kyo, M. et al. Unique patterns of lower respiratory tract microbiota are associated with inflammation and hospital mortality in acute respiratory distress syndrome. Respir. Res. 20, 246 (2019).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
McDonald, D. et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience 1, 7 (2012).
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Hafenbrädl, S., Waeger, D., Marewski, J. N. & Gigerenzer, G. Applied decision making with fast-and-frugal heuristics. J. Appl. Res. Mem. Cogn. 5, 215–231 (2016).
Raab, M. & Gigerenzer, G. The power of simplicity: a fast-and-frugal heuristics approach to performance science. Front. Psychol. 6, 1672 (2015).
Phillips, N. D., Neth, H., Woike, J. K. & Gaissmaier, W. FFTrees: a toolbox to create, visualize, and evaluate fast-and-frugal decision trees. Judgm. Decis. Mak. 12, 344–368 (2017).
Hothorn, T., Hornik, K., van de Wiel, M. A. & Zeileis, A. Implementing a class of permutation tests: the coin package. J. Stat. Softw. 28, 1–23 (2008).
Acknowledgements
We thank the Genomics and Bioinformatics Core Facility of Nantes (GenoBiRD, Biogenouest) for its technical support and the Biological Resource Center for Biobanking of CHU Nantes (Centre de Ressources Biologiques, Nantes, France; BRIF no. BB-0033-00040). We thank D. Flattres Duchaussoy (CHU Nantes) for administrative assistance. The study was funded by the European Union’s Horizon 2020 research and innovation program under grant agreement no. 847782 (HAP2 project). A.R. and R.D. received grants from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 847782. A.R. received funding from the Agence National de la Recherche (ANR) PROGRAM project and the MSD Avenir grant (Phenomenon project). R.D. was funded by the NIH (grants R01HL144599 and K24HL159247). S.V.L. was funded by the NIH (grants RO1 HL143998, P01 AI148104 and P01 AI128482). G.D.K. received funding from the NIH (grants K23 HL139987 and R03 HL162655) and Karius. B.J.K. received funding from the NIH (grant K23 AI121485). R.P.D. received funding from the NIH (grants R01 HL144599 and K24 HL159247) and Horizon H2020 (grant no. 847782). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
E.M., G.D.K., R.P.D. and A.R. selected studies, performed data analyses, interpreted results and drafted the manuscript. J.E.R., Q.L.B., B.J.K., A.P., S.L. and C.S.C. interpreted results and revised the manuscript. All authors have approved the final manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
A.R. has received grants from MSD and bioMerieux, and consulting fees from MSD. G.D.K. has received funding from Karius. S.V.L. is a co-founder and board member of Siolta Therapeutics and has received consulting fees from Sanofi. All other authors have no competing interests.
Peer review
Peer review information
Nature Medicine thanks Leo Celi, Willem Joost Wiersinga and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editor: Alison Farrell, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Consortium members, Supplementary Figures 1–5, Supplementary Tables 1–13, Study protocol.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Montassier, E., Kitsios, G.D., Radder, J.E. et al. Robust airway microbiome signatures in acute respiratory failure and hospital-acquired pneumonia. Nat Med 29, 2793–2804 (2023). https://doi.org/10.1038/s41591-023-02617-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-023-02617-9