Abstract
Artificial intelligence as a medical device is increasingly being applied to healthcare for diagnosis, risk stratification and resource allocation. However, a growing body of evidence has highlighted the risk of algorithmic bias, which may perpetuate existing health inequity. This problem arises in part because of systemic inequalities in dataset curation, unequal opportunity to participate in research and inequalities of access. This study aims to explore existing standards, frameworks and best practices for ensuring adequate data diversity in health datasets. Exploring the body of existing literature and expert views is an important step towards the development of consensus-based guidelines. The study comprises two parts: a systematic review of existing standards, frameworks and best practices for healthcare datasets; and a survey and thematic analysis of stakeholder views of bias, health equity and best practices for artificial intelligence as a medical device. We found that the need for dataset diversity was well described in literature, and experts generally favored the development of a robust set of guidelines, but there were mixed views about how these could be implemented practically. The outputs of this study will be used to inform the development of standards for transparency of data diversity in health datasets (the STANDING Together initiative).
Similar content being viewed by others
Main
Recent years have seen a rapid rise in the development of artificial intelligence (AI) systems for use in healthcare, including those that qualify as a medical device (known as AI as a medical device, AIaMD). This has been enabled by increasing use of electronic health records, accompanied by curation of large-scale health datasets1. However, there are credible concerns that many datasets inadequately reflect the diversity of the individuals or groups contained in the population they are intended to represent. This has previously been described as ‘Health Data Poverty’: a phenomenon where individuals or groups who are underrepresented in health datasets are less able to benefit from data-driven innovations developed using these datasets, including AIaMD2. There is a growing concern that non-diverse and non-representative data contribute to the creation of biased algorithms, resulting in less accurate performance in certain patient groups. Therefore, it is well-recognized that an essential component of ensuring algorithmic safety is to guarantee that datasets are appropriately diverse and representative of their intended use population3.
Data diversity, as measured by equal or relative representation alone, is not enough to achieve equitable outcomes. Even when individuals are represented proportionally in datasets, other forms of bias may be inherently embedded in the representation of those individuals’ data. For example, a dataset may include a proportion of individuals from an ethnic group that is in keeping with national census data (adequate numerical representation), but included individuals from this ethnic group could have a systematically higher likelihood of being misdiagnosed than in the sampled population, meaning the insights derived from this data can remain biased. Although a principal focus of our work is data diversity, we also advocate for a broader view of representativeness in health data, including awareness of the limitations of data collection, data accuracy and ethical concerns around the use of data in minoritized and underserved groups.
Reasons for underrepresentation in datasets broadly fall into two categories: factors that cause individuals or groups to be absent from datasets and factors that cause individuals to be incorrectly or inappropriately categorized into groups despite being present (for example, categories of ‘mixed ethnicity’ or ‘other’). Root causes may include structural barriers to receiving healthcare; barriers to the capture or digitization of relevant health data; individual and structural barriers reducing consent for data sharing; data aggregation, redaction or recoding; collecting data with insufficient granularity; and legal or ethical restrictions on data sharing preventing data accessibility (Fig. 1)2,4,5. The composition and diversity of teams involved in AIaMD development is also critical—teams should include people from different backgrounds as well as those with lived experience of the use case (for example, patients and the public).
Barriers may be present that prevent data about entire groups of people from being included in the dataset (A). They may include barriers to accessing health or social care (meaning data are not generated), inadvertent or deliberate exclusion by the dataset curators or absence of electronic health records (meaning data are not digitized). Certain individuals may be less likely to enter datasets (B); for example, when individuals choose not to allow their data to be included, when methods for data collection are exclusionary (for instance, forcing a binary choice of ‘male’ or ‘female’ for gender), or when redaction occurs after data collection because of legal or ethical restrictions on data sharing. Data may not be collected in sufficient detail, leading to data loss (C; for instance, capturing age in categorical bands such as 20–29 rather than as a continuous variable). Groups of individuals with distinct personal attributes may be merged into a different group either at the point of data collection or by preprocessing after collection (D; for instance, requiring ethnicity or race to be selected from a small list of choices during data capture or combining ethnicity or race groupings into a larger, aggregate group after data capture).
Examples of this lack of diversity in datasets have been previously highlighted in several health areas, including radiology, ophthalmology and dermatology6,7,8. There are further concerns that models may encode biases relating to demographic characteristics even when they are not explicitly trained to do so. This leads to the potential for ‘unknown’ biases reflected in health datasets to become unknowingly and unintentionally embedded in models derived from them9,10.
Despite widespread acknowledgement that inclusiveness is a core tenet of ethical AI in healthcare11,12,13, there remains a shortage of guidance on how to apply such principles in the curation, aggregation and use of health data. The issue of producing data relating to healthcare disparities has previously been explored, with recommendations for data-collection practices, but there are novel challenges in the specific context of AI research14,15. Generic guidelines exist for the improvement of datasets, including a view to reduce healthcare inequalities by promoting patient voice, accurate variables and data linkage14. A commonly raised concern is the reporting of race/ethnicity data, which is variably collected with fragmented and diverse data-collection practices16. The US Food and Drug Administration (FDA) asks for demographic information and inclusion criteria for data collection to be provided if available but does not mandate certain levels of representation across demographic groups in datasets used in AIaMD17. This is a concern particularly pertinent to the field of AIaMD because of the risk of systemic algorithmic bias if models are trained on biased training datasets. AIaMD algorithms learn patterns in the training data and use this to generate predictions when applied to new data. If the data used for training an algorithm are biased against particular demographic groups, the algorithm is likely to underperform when applied to those groups in the real world. Beyond algorithmic bias, diversity in datasets has wider benefits in improving algorithmic performance. A diverse dataset helps AIaMD models generalize their learnings to new and unseen cases. Without diversity, models may perform well on common cases but struggle with unusual or underrepresented ones.
This Analysis aims to explore existing standards, frameworks and best practices that improve data diversity in health datasets in the context of AIaMD. It comprises two parts: (1) a systematic review of the published literature for existing standards, frameworks and best practices; and (2) a survey of stakeholder views to understand how issues of bias and health equity are tackled at present for AIaMD and how best practices can be promoted in the future. This work is part of the STANDING Together initiative (standards for data diversity, inclusivity and generalizability), a program that seeks to develop consensus-driven standards for health data to promote health equity; further information is available at www.datadiversity.org (ref. 18).
Results
Systematic review
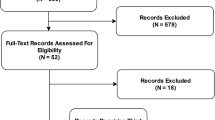
Database searches yielded 10,646 unique records, of which 100 remained after title and abstract screening (Fig. 2). Most of the 10,646 records that were screened did not meet the inclusion criteria, addressing neither health equity nor AIaMD. A further 35 records were screened after identification through reference lists. We identified seven arXiv preprints for the analysis through this method. After full-text screening, 30 relevant records were included.
Of these 30 records, 17 were identified from bibliographic databases, 9 from searches of reference lists and 4 from searches of unindexed conference proceedings (Table 1). All 30 were published between July 2015 and February 2022. Of the 30 records, 1 was published in 2015, 1 in 2017 and 28 since 2018, showing acceleration in the rate of academic discussion around this topic. Most records were authored by individuals from the same department in one institution (14 of 30; 47%), with only 4 of 30 (13%) authored by interdisciplinary, international teams19,20,21,22. Twenty of 30 records (67%) were found in journals, 7 of 30 (23%) were found as preprints and 4 of 30 (13%) were found in conference proceedings. Twenty-seven of 30 (90%) were available open access.
Themes from the literature review
Data were extracted from all 30 records to derive key themes (Extended Data Table 1). Transparency around data-collection practices was a major theme (n = 13), with particular focus on the need for clarity about how data were sampled and for what purpose, how demographic categories were assigned and details of any preprocessing of data. Some articles highlighted the importance of reporting existing health inequalities affecting those included in a dataset, allowing data users to take steps to avoid exacerbating these23,24. The motivation for collecting the data and whether informed consent was obtained from participants was discussed by 11 articles. Furthermore, as legal requirements vary across jurisdictions, there was little consensus about whether or how consent should be obtained from subjects in datasets. Finally, two records discussed how data quality can be improved by involving clinical experts when developing the data-collection strategy5,25.
Missing data were discussed in 10 of 30 records (Extended Data Table 2). Transparency about the causes, extent and consequences of missing data is encouraged, as is transparency about any steps taken to address them. It is understood that increased information about the amount of missing data in a dataset will promote transparency and considerations about appropriateness of use, given that usefulness and generalizability of AI and machine learning (ML) models are impaired by missing data19. Research into missing data also revealed a recommendation that aggregation of demographic groups or variables into a smaller number of groups should be reported, which would similarly help researchers understand the limitations of the dataset26.
Data labeling was addressed by 8 of 30 records (Extended Data Table 3). Transparency was again an overarching theme: articles discuss the need to report how labeling was performed—particularly whether labels represent ground truths and whether there are known or potential biases in labels, such as if they were reported by humans (and therefore subject to inter-reporter variability). One article encouraged analysis of statistical relationships between labels and demographic factors so that potential confounders can be identified and controlled for during model development5.
A recurring theme was the identification of groups at risk of harm. These groups were variably described by authors as “vulnerable”, “minority”, “minoritized”, “underserved”, “marginalized” and “protected”. The cross-cutting theme was that these are groups considered to be more susceptible to physical, social or economic harm. The issue of certain groups being at greater risk of vulnerability was discussed in 24 of 30 records (Extended Data Table 4). However, there was little consensus about how biases should be addressed or which groups are most at risk. Suggested approaches to identify and reduce bias and harms for demographic subgroups included predefining groups suspected to be at risk, targeting data collection and model development to benefit these groups in particular, ensuring that representatives from at-risk groups are involved with model development (including as experts in a development team, such as developers, programmers and analysts), testing data for confounders rather than automatically including all features in training data and testing model performance in minoritized subgroups. Attributes specified as being particularly at risk of harm with underrepresentation include ethnicity, race, pregnancy status, age, nationality, gender, sex, socioeconomic status, religion, indigenous and tribal community membership, disability status, sexual orientation, preferred language, Fitzpatrick skin type, health status, education, employment status, geographical location and marital status.
Stakeholder survey
Whereas the systematic review provided an oversight of current best-practice principles for health datasets in AI, the stakeholder survey provided insights into how principles could be operationalized and by whom. Twenty participants completed the scoping survey. Of these participants, ten (50%) reported their sex as female, nine (45%) reported their sex as male, and one (5%) did not provide this information. Eighteen participants (90%) reported that their gender identity was the same as the sex registered at birth, one participant (5%) reported that their gender identity was different than their sex registered at birth and one participant (5%) did not provide this information. Four main themes and 17 subthemes were identified (Table 2).
The first theme was ‘the role of demographic data’. Stakeholders used demographic data in several ways to assess the safety and efficacy of AIaMDs across different subgroup populations. Ensuring that representative data are used to train and validate AIaMDs for the population in which they are to be deployed was felt to be most important. Ways of demonstrating representativeness included describing the intended use and users of the AIaMD, identifying subgroups of interest up front and being transparent about poor performance. Race and ethnicity data were seen as an important means to explore known and unknown biases potentially leading to health inequalities.
“Models must be able to ‘work’ for those belonging to racialised minority groups, and clinicians/researchers/developers must go through stringent governance measures to ensure inequalities, racism and other forms of discrimination are not exacerbated by use of medical AI models.”
Challenges of ensuring diversity in datasets included issues of lack of health data in certain populations (health data poverty), lack of standardization across attribute categories, difficulty in harmonizing several methods of data capture and data-governance restrictions. Other factors relating to the development pathway included poorly defined use cases, a lack of relevant stakeholder input, difficulty in accessing suitable datasets and existing gaps in current evidence about underserved populations at risk of harm linked to health outcomes. Stakeholders used various solutions to derive missing demographic data, including statistical techniques such as imputation, Bayesian geocoding and linking across several datasets. Stakeholders suggested they would like to see policy changes, standards of best practice with a statement of scope up front describing data diversity and regulatory authorities providing clarity about specific requirements according to the intended use, as well as toolkits to tackle health data poverty issues.
“Assist various bodies in addressing data poverty (I see this as the core problem that needs solving).”
“Using linked data records and collating ethnicity information from different parts of the healthcare record can improve ethnicity data capture.”
“Multiple imputations can be helpful for some variables but is sub-optimal. There are weak surrogates/indirect methods of increasing populating ethnicity variables.”
The second theme was ‘data diversity’. The definition of dataset diversity was generally considered a universal concept whose application should be context specific depending on the research question or intended use population. The scope of data diversity should be broad and include race, ethnicity, age, gender, sex, socioeconomic status, clinically relevant disease populations, neurodiversity, disability, language barriers and educational level. It was felt that an ideal dataset should represent a global population that is diverse enough to enable a range of problems to be explored with adequate statistical power. Race and ethnicity were seen as dataset attributes that ‘must be included’. Barriers identified included lack of standardization across the globe, low statistical power in underrepresented groups/rare events and a lack of knowledge about intended use populations/subgroups and their health outcomes. Although race and ethnicity are nearly universally acknowledged as important, there is considerable lack of precision and understanding of the terms in science more broadly27.
“Diversity as a concept is universal, but its application is contextual.”
“Diversity in health datasets should be used more broadly than simply protected characteristics such as sex and ethnicity.”
The third theme was ‘the use of metrics’. In principle, most respondents were in favor of some type of metric to measure diversity but unsure about how this could be operationalized. There was a definite lack of consensus about the concept of introducing a rating for the diversity of a dataset, with considerable concerns about how a rating of diversity could be implemented. When metrics were considered, stakeholders felt they would have greater value if externally validated rather than solely reported by the dataset’s curators. An alternative approach was that dataset curators could be validated as ‘safe providers’, meaning curators and/or their organizations would demonstrate adherence to standards across all datasets they produce. Other metrics put forward included level of inclusion, completeness and/or missingness of demographic data and distribution of the data. Interestingly, participants also proposed metrics that were related indirectly to the data itself but rather to the model derived. For example, participants discussed measures of model performance (such as systematic error rates across subgroups) that could result from biases in the data. However, respondents anticipated many challenges, such as knowing up front for which subpopulations poor performance should be specifically tested, a lack of established methods for evaluating performance and comparability of variables across different datasets.
“I am not sure this could be distilled into a simple set of metrics but rather see a minimum requirement of descriptive information re: a dataset as an option that is proportion of different ethnicities, age groups, gender.”
“I don’t think datasets should be ‘rated’ on demographic diversity personally. To me, this borders on saying some are ‘better’ than others but it is context-specific and depends on the planned setting of deployment.”
The fourth theme was ‘standards’. There was consensus about the importance of standards to enable risks and harms to be identified, improve the quality of the datasets, address bias and provide accountability. However, respondents cautioned against imposing strict diversity requirements, as doing so might risk products being withdrawn for already-marginalized populations, as well as unethical data-gathering practices28. Recommendations for adoption included making the standards part of the procurement, funding or product-approval process; making them part of the publishing pathway; or implementing an accreditation pathway for organizations to demonstrate compliance. However, there was no consensus about which organizations should be responsible for mandating compliance.
“It’s great that the issue has been recognised, but more needs to be done to change things, for example by having a standard for researchers to consider and apply when designing their studies or curating new datasets, and developing methods to ensure people are held accountable.”
Regarding whether such standards should be mandated, some respondents suggested that mandating the standards through regulators, journal editors or commissioners was an acceptable approach, provided diversity of the populations in the dataset was the requirement. If diversity across populations related to performance of the model was to be mandated for compliance, this approach risked setting a level that could not be achieved by creating a problem with enforcement and increasing costs. The consequences might include slowing or even stifling research and innovation because of data poverty issues. Some felt that a softer approach was needed, such as defining best practice with a set of data diversity standards and encouraging adoption and self-reporting. Whichever enforcement mechanism is implemented, practical tools can be developed to improve uptake, such as providing guidance on how to adhere to the standards, making validation datasets available, engagement with stakeholders, providing incentives and removing barriers. The greatest barriers to adherence identified were resources such as time and cost.
“Funders at the proposal stage, journals at the publication stage and in parallel the regulatory system.”
“If you propose a body for checking compliance more formally, you will need to fund them as well. However, if the stakeholders who are required to comply with the standards feel there is just one more thing being added to their increasing requirements, you may risk facing a rebellion unless this work is adequately compensated. I suggest soft standards rather than formal checks.”
Discussion
This study used two methods: a systematic review and a scoping survey of expert stakeholders. The systematic review identified a range of recommendations related to data diversity, with the scoping survey supplementing it by exploring practical considerations relating to their implementation. To our knowledge, this is the first systematic synthesis of existing standards, frameworks, guidelines and best practices for the use of health datasets in the context of AIaMD. We found a clear consensus that there is a need for diversity in datasets and that issues of algorithmic bias may prevail where this is absent. The increasing resources directed towards creating large-scale health datasets has not been accompanied by equivalent efforts to ensure that they are adequately representative and diverse. Even when guidance recognizes the importance of data diversity, it is high-level and not easily operationalized.
The systematic review provided insight into existing guidelines for data collection, handling missing data and labeling data. A key theme found through the systematic review was a need for transparency in how datasets are prepared, including who is included or excluded from the dataset, how missing data are handled and how data are labeled. Greater transparency in these areas allows better understanding of the context and limitations of a dataset, which in turn provides a guide to the potential limitations of any inferences or innovations derived from that dataset. For example, if a dataset excludes certain groups, this information should be evident to potential users of the dataset and should be reported alongside insights derived from that dataset to provide context and the likely scope of application.
With regard to personal attributes, among all included articles, sex, age, race and ethnicity were most commonly cited as attributes that could associate a group with risk of harm or disadvantage; however, there was a notable lack of literature addressing how these concerns could be addressed for these groups. Less commonly, records cited pregnancy, income, marital status and a range of other attributes that may require further consideration. There was notable overlap between attributes identified as often lacking diversity and those that are specifically protected by various jurisdictions, such as in the UK Equality Act 2010, the European Union Charter of Fundamental Rights and the United Nations29,30,31. The importance of collecting demographic attribute information is not necessarily for it to be embedded as an input variable or predictor in ML models (which may perpetuate harm along these axes) but importantly, to ensure that it is collected for auditing model performance across disaggregated subgroups as a method of bias discovery. In our literature review, ‘Healthsheet’ and ‘Datasheets for Datasets’ were the most comprehensive guides to data documentation, with elements in both relating to diversity32,33.
There was a high degree of concordance between the recommendations gathered from the systematic review and those obtained through our survey. However, the scoping survey highlighted the potential difficulties and lack of pragmatic guidance about how such guidelines can be implemented practically as well as who should be responsible for overseeing their implementation. This indicates that the issues of interest are well-recognized and conceptually understood; however, there is a clear need to focus on operationalizing existing knowledge. The scoping survey additionally identified barriers and potential enablers to creating standards and best practices that are ready for translation into the real world. It highlighted that although standards would be beneficial, there must be some means of implementing them, which could include embedding them into a product-approval process or making them prerequisites for eligibility for funding, health technology appraisal or procurement. Standards could also be implemented by externally provided accreditation, which may be seen favorably by funders, regulatory bodies and research boards. Another option includes voluntary self-reporting of standards with minimal external oversight.
It was also highlighted that it is not clear which agencies could mandate or suggest completion of such standards. Journal editors, regulators, health policy organizations and funders were among the suggested agencies. In health research, the Enhancing the Quality and Transparency of Health Research (EQUATOR) network is well-recognized as a resource for gold-standard guidance for study reporting, endorsing widely used guidelines including Consolidated Standards of Reporting Trials 2010 guidelines for randomized controlled trials. Although the network focuses on reporting and transparency of research studies, a similar structure may be considered for datasets. The role of regulatory agencies in mandating the data diversity used in AIaMD is unclear, as their remit is at present limited to ensuring that datasets reflect the intended use population. Although respondents acknowledged the lack of evidence on this topic, a counterargument to heavy-handed enforcement was also raised from the stakeholder survey, with the perception that enforcing higher expectations for datasets may stifle innovation, impede health improvement and possibly exacerbate inequalities as a result. Overall, the findings of the survey indicated strong support for the development of standards but ambiguity as to their implementation.
This study involved a comprehensive review of academic literature reporting recommendations for dataset use, but it did not extend to reviewing gray literature, including governmental reports. However, we have noted that the issue of algorithmic bias has been formally recognized by the UK Medicines and Healthcare products Regulatory Agency, UK government, the UK Information Commissioner’s Office, the US FDA and the European Parliament34,35,36,37,38,39. “Ensuring inclusiveness and equity” is one of six principles for AI development prescribed by the World Health Organization, referring to “age, sex, gender, income, race, ethnicity, sexual orientation, ability or other characteristics protected under human rights codes”13. The same WHO report states, “No technology, AI or otherwise, should sustain or worsen existing forms of bias and discrimination”13.
The search was limited to records published since 2015. This is a limitation to the study, but it is noted that the results exhibited an exponential increase in the number of relevant records published since 2018. Only one relevant record was published in 2015. It would be expected that any notable pre-2015 papers should have been revealed in reference-list screening, but none were. Accordingly, setting an earlier start date for screening would have been unlikely to identify any more relevant articles and would have substantially increased the number of papers it was necessary to screen. Data collection was performed by two independent reviewers for 11 of the records. Data for the remaining articles were extracted by a single reviewer, who extracted data from all papers included in the analysis for consistency of voice in the extracted summaries. Although data collection by a single author is a limitation, use of a standardized data-extraction sheet partially mitigates it.
This study has focused on factors that encourage dataset diversification as a lever to address health data poverty. Underrepresentation of minority groups in datasets is well-recognized as an important driver of algorithmic bias, but other mechanisms can be applied downstream to mitigate its effects. This includes practicing model diversity: for example, producing several models and combining the outputs with ensemble learning to diversify the parameters considered by the model and reduce the risk of overfitting to an unbalanced data sample. Synthetic data have also been recently proposed as a method of selectively generating data for marginalized populations to rebalance datasets; however, this approach has limitations in that it effectively oversamples from a small group without truly gaining diversity40,41,42. It should be emphasized that these (and other) methods are an active area of research with a need for empirical evidence to prove their applicability. Similarly, this review does not extend into other ethical implications of the use of ML in healthcare, although this topic has been extensively studied previously and includes other issues of privacy, trust and accountability43.
We took a broad approach to the recruitment of different types of stakeholders in the scoping survey and therefore did not apply a formal sampling framework. As a result, we cannot be sure the voices represented are consistent across all stakeholders. This survey was intended as a scoping exercise and not meant to be an exhaustive qualitative study. Future planned work as part of the STANDING Together initiative involves input from a wider gamut of stakeholders including patient and public partners (two of whom are coauthors of this study).
The issue of algorithmic bias is well described in medical literature, with dataset insufficiency a key driver. Although principles such as Findable, Accessible, Interoperable, and Reusable (FAIR) seek to improve data availability and use more generally, there is a paucity of data for certain groups and lack of diversity in existing datasets44. This Analysis has highlighted the importance of curating and aggregating health data to promote diversity, inclusivity and equity as well as the lack of guidelines available to facilitate doing so. Although reporting guidelines exist for randomized controlled trials using AI, they focus on the reporting of study results rather than the design and use of datasets45. Future avenues of research may seek to produce clear guidelines for the development and use of datasets, revolving around the need for diversity and inclusion of marginalized populations and improving data interoperability by means of common data models and standards such as Observational Medical Outcomes Partnership (OMOP) and Fast Healthcare Interoperability Resources (FHIR). Specific guidance about new development practices (including use of synthetic data, federated learning and foundation models) is also needed.
Transparent documentation around diversity and appropriateness of datasets used in AIaMD development will help commissioners, clinicians and health systems determine the risk of bias so they can make informed decisions around whether to deploy corresponding algorithms for their population. The STANDING Together project (https://www.datadiversity.org/) is one such endeavor. Building on the outputs of this systematic review and stakeholder survey, STANDING Together is developing consensus-derived standards coauthored by an international, interdisciplinary team that reinforce ethics and inclusivity in the documentation and use of healthcare datasets, allowing developers to ensure that AIaMD works for everyone18. The findings described in this literature review and stakeholder survey will directly inform the proposed items for STANDING Together, and the methods describing their translation into specific, actionable recommendations will be outlined in a subsequent paper. The recommendations will undergo a multistakeholder, three-staged Delphi study, and the resulting standards will be available in late 2023.
Methods
This research was conducted in compliance with all relevant ethical regulations, including informed consent from all participants. Ethical approval was granted by the University of Birmingham’s Science, Technology, Engineering and Mathematics Ethical Review Committee (ERN_21-1831).
Systematic review
We searched for records describing existing standards, frameworks and best practices for ensuring data diversity in health datasets in the context of AIaMD. An informatician was consulted for the development of the search strategy (Systematic Review Search Strategy), and the searches were conducted on 10 October 2021 on MEDLINE (Ovid), Embase (Ovid) and Web of Science. Results were limited to publications since 2015 in the English language. E-publications ahead of print, in-process publications, in-data-review and other non-indexed citations were included through the MEDLINE search. Deduplication was carried out in EndNote 20 (Clarivate, 2013) and screening in Covidence (https://www.covidence.org/; Veritas Health Innovation, 2022). It was recognized that relevant results may exist as preprints that may not be covered by traditional systematic review searches. As preprint databases are not typically covered by searchable databases, nor are their websites conducive to systematic searches, the burden of the workload to replicate our strategy on the medRxiv and arXiv engines would have been unmanageable. MedRxiv is indexed by Embase, but arXiv is not indexed by any of the databases used. To mitigate this limitation, we conducted reference-list screening for all 100 articles included for full-text screening. Reference lists of included records were searched to identify relevant preprints (including arXiv and medRxiv) or other potentially relevant records. Further searches were conducted of the past five years’ worth of archives of relevant conference proceedings for ML and AI in healthcare: ‘Machine Learning for Health’ (https://ml4health.github.io/2022/), ‘Machine Learning for Healthcare’ (https://www.mlforhc.org) and ‘Conference on Health, Inference, and Learning’ (https://www.chilconference.org/). This scoping review was conducted following the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement, and a PRISMA flow chart was also created (Fig. 2)64. Abstract screening was performed by two authors independently (A.A., J.A. or X.L.). Non-consensus was resolved by discussion and involvement of a third reviewer if necessary. Before full-text screening, 10% of the records were full-text screened independently by two reviewers as part of a pilot. Once a high degree of concordance was reached between two independent reviewers, the remaining records were assessed by a single reviewer. Studies were included if they presented standards, frameworks or guidance for AI or health data about issues intersecting AI and bias, fairness, health equity and representation, and coding/categorization of minoritized and marginalized groups. Exclusion criteria were guidance not related to health data, guidance relating to technical and infrastructural aspects of health dataset curation only, guidance relating to privacy and governance and cybersecurity only.
For each record, a single reviewer extracted data using a predefined data-extraction sheet. Bibliometric information about each record was extracted, including publication date, number of citations and details of the authorship team (in terms of single/several institutions, geographies and disciplinary background). If specific vulnerable or minority patient groups were discussed in the record, they were also recorded separately. When it was reported, we extracted information about the methodology that led to the construction of the recommendations, including descriptions of any literature review and stakeholder involvement.
Stakeholder survey
We approached 45 participants representing individuals who work in health-data research and/or AIaMD, including dataset curators, academics, clinicians and medical-device regulators. Participants were identified as authors of relevant publications and through consultation with the STANDING Together working group on the basis of expertise and previous work in healthy inequity, medical datasets and AIaMD. Respondents were invited by email to participate in the survey using Qualtrics (Qualtrics XM, 2018; https://www.qualtrics.com).
The survey consisted of 14 free-text questions (Supplementary File 5) exploring how issues of bias and risk of health inequity are at present tackled for AIaMD and how best-practice recommendations could be operationalized. For data analysis, we took an inductive thematic analysis approach that was exploratory and descriptive in nature65. One author (J.P.) conducted the analysis and used several iterations of the responses to the survey questions to refine the initial list of codes and create a codebook using NVivo (NVivo release 1.0, March 2020, QSR International; https://lumivero.com/products/nvivo/). Two authors (E.L. and X.L.) reviewed the data extracts and relevant codes independently, and then all three of the above authors discussed and agreed on the final codebook.
Statistics and reproducibility
Data relating to the effect of articles included in our systematic review (including journal impact factor, citation count and altmetric data) were obtained but not included in the analysis because these data were not necessary to extract themes from the included articles. No statistical method was used to predetermine the sample size for the stakeholder survey. Blinding and randomization are not applicable to non-interventional studies.
Systematic review search strategy
MEDLINE search strategy
Ovid MEDLINE(R) and Epub Ahead of Print, In-Process, In-Data-Review & Other Non-Indexed Citations, Daily and Versions(R) <1946 to October 08, 2021>
-
1.
cultural diversity/ 12175
-
2.
(ethic* or divers* or fairness or fair or bias or biased or pluralism* or multicultural*).mp. [mp=title, abstract, original title, name of substance word, subject heading word, floating subheading word, keyword heading word, organism supplementary concept word, protocol supplementary concept word, rare disease supplementary concept word, unique identifier, synonyms] 1124521
-
3.
1 or 2 1124521
-
4.
exp Artificial Intelligence/ 124778
-
5.
(Artificial intelligence or AI or Natural Language processing or NLP or Machine learning or Support Vector Machine* or neural network* or deep learning).mp. [mp=title, abstract, original title, name of substance word, subject heading word, floating sub-heading word, keyword heading word, organism supplementary concept word, protocol supplementary concept word, rare disease supplementary concept word, unique identifier, synonyms] 196657
-
6.
((health or patient or medical) adj2 (data* or record*)).mp. [mp=title, abstract, original title, name of substance word, subject heading word, floating sub-heading word, keyword heading word, organism supplementary concept word, protocol supplementary concept word, rare disease supplementary concept word, unique identifier, synonyms] 324576
-
7.
exp Medical Records/ 152836
-
8.
4 or 5 or 6 or 7 587250
-
9.
guideline/ 16447
-
10.
(standard* or guidance or guideline* or framework* or policy or policies or governance).mp. [mp=title, abstract, original title, name of substance word, subject heading word, floating sub-heading word, keyword heading word, organism supplementary concept word, protocol supplementary concept word, rare disease supplementary concept word, unique identifier, synonyms] 3063858
-
11.
9 or 10 3063858
-
12.
3 and 8 and 11 9897
-
13.
limit 12 to yr=“2015 -Current” 6089
-
14.
limit 13 to lg=“english” 5995
Embase search strategy
Embase <1996 to 2021 Week 40>
-
1.
cultural diversity/ 2415
-
2.
(ethic* or divers* or fairness or fair or bias or biased or pluralism* or multicultural*).mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword heading word, floating subheading word, candidate term word] 1235079
-
3.
1 or 2 1235079
-
4.
exp Artificial Intelligence/ 51842
-
5.
(Artificial intelligence or AI or Natural Language processing or NLP or Machine Learning or Support Vector Machine* or neural network* or deep learning).mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword heading word, floating subheading word, candidate term word] 237951
-
6.
((health or patient or medical) adj2 (data* or record*)).mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword heading word, floating subheading word, candidate term word] 582323
-
7.
exp medical record/ 255321
-
8.
4 or 5 or 6 or 7 822925
-
9.
(standard* or guidance or guideline* or framework* or policy or policies or governance).mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword heading word, floating subheading word, candidate term word] 3535573
-
10.
3 and 8 and 9 13849
-
11.
limit 10 to yr=“2015 -Current” 8994
-
12.
limit 11 to lg=“english” 8925
Web of Science strategy
((((((((TS=(“ethic*”)) OR TS=(“divers*”)) OR TS=(“fairness”)) OR TS=(“fair”)) OR TS=(“bias”)) OR TS=(“biased”)) OR TS=(“pluralism*”)) OR TS=(“multicultural*”)) AND ((((((((TS=(“Artificial Intelligence”)) OR TS=(“AI”)) OR TS=(“Natural Language processing”)) OR TS=(“NLP*”)) OR TS=(“Machine Learning”)) OR TS=(“Support Vector Machine*”)) OR TS=(“neural network*”)) OR TS=(“deep learning”)) AND TS=((“health” OR “patient” OR “medical”) NEAR/2 (“data*” OR “record*”)) AND (((((((TS=(“standard*”)) OR TS=(“guidance”)) OR TS=(“guideline*”)) OR TS=(“framework*”)) OR TS=(“policy”)) OR TS=(“policies”)) OR TS=(“governance”))
Refined to results from 2015
Refined to results in English language
307 results
Reporting summary
Further information on the research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All relevant data are included in the manuscript and supplementary files. Reproducible searches for Web of Science (https://webofscience.com/), Ovid MEDLINE (through ovid.com) and Embase (through ovid.com) are also included in the Methods, with relevant direct links.
Code availability
No computer code was used. Qualtrics XM (https://www.qualtrics.com/uk/) was used to support the survey. NVivo was used to help refine the initial list of codes from the survey to create a codebook (https://www.qsrinternational.com/nvivo-qualitative-data-analysis-software/home).
References
Sidey-Gibbons, J. A. M. & Sidey-Gibbons, C. J. Machine learning in medicine: a practical introduction. BMC Med. Res. Methodol. 19, 64 (2019).
Ibrahim, H., Liu, X., Zariffa, N., Morris, A. D. & Denniston, A. K. Health data poverty: an assailable barrier to equitable digital health care. Lancet Digit. Health 3, e260–e265 (2021).
Kuhlman, C., Jackson, L. & Chunara, R. No computation without representation: avoiding data and algorithm biases through diversity. In Proc. 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining (KDD '20) 3593 (ACM, 2020); https://doi.org/10.1145/3394486.3411074
Courbier, S., Dimond, R. & Bros-Facer, V. Share and protect our health data: an evidence based approach to rare disease patients’ perspectives on data sharing and data protection - quantitative survey and recommendations. Orphanet J. Rare Dis. 14, 175 (2019).
Chen, I. Y. et al. Ethical machine learning in healthcare. Annu Rev. Biomed. Data Sci. 4, 123–44. (2021).
Khan, S. M. et al. A global review of publicly available datasets for ophthalmological imaging: barriers to access, usability, and generalisability. Lancet Digit. Health 3, e51–e66 (2021).
Wen, D. et al. Characteristics of publicly available skin cancer image datasets: a systematic review. Lancet Digit. Health 4, e64–e74 (2022).
Kaushal, A., Altman, R. & Langlotz, C. Geographic distribution of US cohorts used to train deep learning algorithms. JAMA 324, 1212–1213 (2020).
Gichoya, J. W. et al. AI recognition of patient race in medical imaging: a modelling study. Lancet Digit. Health 4, e406–e414 (2022).
Glocker, B., Jones, C., Bernhardt, M. & Winzeck, S. Risk of bias in chest radiography foundation models. Radiol. Artif. Intell. https://doi.org/10.1148/ryai.230060 (2023).
Zou, J. & Schiebinger, L. Ensuring that biomedical AI benefits diverse populations. eBioMedicine https://doi.org/10.1016/j.ebiom.2021.103358 (2021).
Jobin, A., Ienca, M. & Vayena, E. The global landscape of AI ethics guidelines. Nat. Mach. Intell. 1, 389–399. (2019).
Ethics and Governance of Artificial Intelligence for Health (WHO 2021); https://www.who.int/publications-detail-redirect/9789240029200
Block, R. G. et al. Recommendations for improving national clinical datasets for health equity research. J. Am. Med. Inform. Assoc. 27, 1802–1807 (2020).
DeVoe, J. E. et al. The ADVANCE network: accelerating data value across a national community health center network. J. Am. Med. Inform. Assoc. 21, 591–595 (2014).
Hasnain-Wynia, R. & Baker, D. W. Obtaining data on patient race, ethnicity, and primary language in health care organizations: current challenges and proposed solutions. Health Serv. Res. 411, 1501–1518 (2006).
Computer-Assisted Detection Devices Applied to Radiology Images and Radiology Device Data - Premarket Notification [510(k)] Submissions. (FDA, 2022); https://www.fda.gov/regulatory-information/search-fda-guidance-documents/computer-assisted-detection-devices-applied-radiology-images-and-radiology-device-data-premarket
Ganapathi, S. et al. Tackling bias in AI health datasets through the STANDING Together initiative. Nat. Med. 28, 2232–2233 (2022).
Vollmer, S. et al. Machine learning and artificial intelligence research for patient benefit: 20 critical questions on transparency, replicability, ethics, and effectiveness. Br. Med. J. 368, l6927 (2020).
Challen, R. et al. Artificial intelligence, bias and clinical safety. BMJ Qual. Saf. 28, 231–237 (2019).
Wiens, J. et al. Do no harm: a roadmap for responsible machine learning for health care. Nat. Med. 25, 1337–40. (2019).
Saleh, S., Boag, W., Erdman, L. & Naumann, T. Clinical collabsheets: 53 questions to guide a clinical collaboration. In Proc. 5th Machine Learning for Healthcare Conference (eds Doshi-Velez, F. et al.) 783–812 (PMLR, 2022); https://proceedings.mlr.press/v126/saleh20a.html
Ferryman, K. Addressing health disparities in the Food and Drug Administration’s artificial intelligence and machine learning regulatory framework. J. Am. Med. Inform. Assoc. 27, 2016–2019 (2020).
Suresh, H. & Guttag, J. A framework for understanding sources of harm throughout the machine learning life cycle. In Proc. Equity and Access in Algorithms, Mechanisms, and Optimization 1–9 (ACM, 2021); https://dl.acm.org/doi/10.1145/3465416.3483305
Lysaght, T., Lim, H. Y., Xafis, V. & Ngiam, K. Y. AI-assisted decision-making in healthcare: the application of an ethics framework for big data in health and research. Asian Bioeth. Rev. 11, 299–314 (2019).
Flanagin, A., Frey, T., Christiansen, S. L. & Bauchner, H. The reporting of race and ethnicity in medical and science journals: comments invited. JAMA 325, 1049–1052 (2021).
Cerdeña, J. P., Grubbs, V. & Non, A. L. Racialising genetic risk: assumptions, realities, and recommendations. Lancet 400, 2147–2154. (2022).
Elias, J. Google contractor reportedly tricked homeless people into face scans. CNBC https://www.cnbc.com/2019/10/03/google-contractor-reportedly-tricked-homeless-people-into-face-scans.html (2019).
Equality Act 2010. Statute Law Database (UK Government, 2010); https://www.legislation.gov.uk/ukpga/2010/15/section/4
Declaration of the High-Level Meeting of the General Assembly on the Rule of Law at the National and International Levels (UN General Assembly, 2012); https://digitallibrary.un.org/record/734369
Article 21 - Non-Discrimination (European Union Agency for Fundamental Rights, 2007); https://fra.europa.eu/en/eu-charter/article/21-non-discrimination
Gebru, T. et al. Datasheets for datasets. Preprint at http://arxiv.org/abs/1803.09010 (2021).
Rostamzadeh, N. et al. Healthsheet: development of a transparency artifact for health datasets. In Proc. 2022 ACM Conference on Fairness, Accountability, and Transparency 1943–1961 (ACM, 2022); https://doi.org/10.1145/3531146.3533239
Smeaton, J. & Christie, L. AI and healthcare. UK Parliament POSTnote https://post.parliament.uk/research-briefings/post-pn-0637/ (2021).
Human bias and discrimination in AI systems. ICO https://webarchive.nationalarchives.gov.uk/ukgwa/20211004162239/https://ico.org.uk/about-the-ico/news-and-events/ai-blog-human-bias-and-discrimination-in-ai-systems/ (2019).
Artificial Intelligence and Machine Learning in Software as a Medical Device (FDA, 2021); https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device
A Governance Framework for Algorithmic Accountability and Transparency (European Parliament, Directorate-General for Parliamentary Research Services, 2019); https://data.europa.eu/doi/10.2861/59990
WHO Issues First Global Report on Artificial Intelligence (AI) in Health and Six Guiding Principles for Its Design and Use (WHO, 2021); https://www.who.int/news/item/28-06-2021-who-issues-first-global-report-on-ai-in-health-and-six-guiding-principles-for-its-design-and-use
Regulatory Horizons Council: The Regulation of Artificial Intelligence as a Medical Device. (UK Government, 2022); https://www.gov.uk/government/publications/regulatory-horizons-council-the-regulation-of-artificial-intelligence-as-a-medical-device
Arora, A. & Arora, A. Generative adversarial networks and synthetic patient data: current challenges and future perspectives. Future Healthc. J. 9, 190–193 (2022).
Burlina, P., Joshi, N., Paul, W., Pacheco, K. D. & Bressler, N. M. Addressing artificial intelligence bias in retinal diagnostics. Transl. Vis. Sci. Technol. 10, 13 (2021).
Koivu, A., Sairanen, M., Airola, A. & Pahikkala, T. Synthetic minority oversampling of vital statistics data with generative adversarial networks. J. Am. Med. Inform. Assoc. 27, 1667–74. (2020).
Murphy, K. et al. Artificial intelligence for good health: a scoping review of the ethics literature. BMC Med. Ethics 22, 14 (2021).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Liu, X., Cruz Rivera, S., Moher, D., Calvert, M. J. & Denniston, A. K. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med. 26, 1364–74. (2020).
Villarroel, N., Davidson, E., Pereyra-Zamora, P., Krasnik, A. & Bhopal, R. S. Heterogeneity/granularity in ethnicity classifications project: the need for refining assessment of health status. Eur. J. Public Health 29, 260–266 (2019).
Denton, E. et al. Bringing the people back in: contesting benchmark machine learning datasets. Preprint at http://arxiv.org/abs/2007.07399 (2020).
Holland, S., Hosny, A., Newman, S., Joseph, J. & Chmielinski, K. The dataset nutrition label: a framework to drive higher data quality standards. Preprint at http://arxiv.org/abs/1805.03677 (2018).
Floridi, L., Cowls, J., King, T. C. & Taddeo, M. How to design AI for social good: seven essential factors. Sci. Eng. Ethics 26, 1771–1796. (2020).
Char, D. S., Abràmoff, M. D. & Feudtner, C. Identifying ethical considerations for machine learning healthcare applications. Am. J. Bioeth. 20, 7–17 (2020).
Griffiths, K. E., Blain, J., Vajdic, C. M. & Jorm, L. Indigenous and tribal peoples data governance in health research: a systematic review. Int. J. Environ. Res. Public Health 18, 10318 (2021).
Hernandez-Boussard, T., Bozkurt, S., Ioannidis, J. P. A. & Shah, N. H. MINIMAR (MINimum Information for Medical AI Reporting): developing reporting standards for artificial intelligence in health care. J. Am. Med. Inform. Assoc. 27, 2011–2015 (2020).
Paulus, J. K. & Kent, D. M. Predictably unequal: understanding and addressing concerns that algorithmic clinical prediction may increase health disparities. NPJ Digit. Med. 3, 1–8 (2020).
McCradden, M. D., Joshi, S., Mazwi, M. & Anderson, J. A. Ethical limitations of algorithmic fairness solutions in health care machine learning. Lancet Digit. Health 2, e221–e223 (2020).
Douglas, M. D., Dawes, D. E., Holden, K. B. & Mack, D. Missed policy opportunities to advance health equity by recording demographic data in electronic health records. Am. J. Public Health 105, S380–S388 (2015).
Mitchell, M. et al. Model cards for model reporting. In Proc. Conference on Fairness, Accountability, and Transparency (FAT* '19) 220–229 (ACM, 2019); https://doi.org/10.1145/3287560.3287596
Mörch, C. M., Gupta, A. & Mishara, B. L. Canada protocol: an ethical checklist for the use of artificial Intelligence in suicide prevention and mental health. Artif. Intell. Med. 108, 101934 (2020).
Saleiro, P. et al. Aequitas: a bias and fairness audit toolkit. Preprint at http://arxiv.org/abs/1811.05577 (2019).
Xafis, V. et al. An ethics framework for big data in health and research. Asian Bioeth. Rev. 11, 227–254. (2019).
Abstracts from the 53rd European Society of Human Genetics (ESHG) conference: e-posters. Eur. J. Hum. Genet. 28, 798–1016 (2020).
Zhang, X. et al. Big data science: opportunities and challenges to address minority health and health disparities in the 21st century. Ethn. Dis. 27, 95–106 (2017).
Rajkomar, A., Hardt, M., Howell, M. D., Corrado, G. & Chin, M. H. Ensuring fairness in machine learning to advance health equity. Ann. Intern. Med. 169, 866–872 (2018).
Fletcher, R. R., Nakeshimana, A. & Olubeko, O. Addressing fairness, bias, and appropriate use of artificial intelligence and machine learning in global health. Front. Artif. Intell. https://doi.org/10.3389/frai.2020.561802 (2021).
The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372, n71 (2021).
Braun, V. & Clarke, V. Using thematic analysis in psychology. Qual. Res. Psychol. 3, 77–101 (2006).
Acknowledgements
This project is funded by the NHS AI Lab at the NHS Transformation Directorate and The Health Foundation and managed by the National Institute for Health and Care Research (AI_HI200014). The views expressed in this publication are those of the author(s) and not necessarily those of the NHS Transformation Directorate, The Health Foundation or the National Institute for Health and Care Research. D.T. and F.M. are funded by the National Pathology Imaging Co-operative, NPIC (project no. 104687), supported by a £50 million investment from the Data to Early Diagnosis and Precision Medicine strand of the government’s Industrial Strategy Challenge Fund, managed and delivered by UK Research and Innovation (UKRI).
Author information
Authors and Affiliations
Contributions
A.A., J.A., J.P., A.D. and X.L. conceived and designed the project. A.D. and X.L. were primarily responsible for the planning and supervision of the project. A.A., J.A. and X.L. were responsible for data collection through literature review. J.A., S.G. and X.L. were responsible for data collection through survey delivery. A.A., J.P., J.A. and X.L. drafted the initial manuscript. S.G., E.L., M.D.M., L.O.-R., S.R.P., M.G., F.M., D.T., N.R., B.M., J.G., A.O.A., S.K., R.M., K.H., E.S., N.J.S., H.C.-L., M.C. and A.D. contributed to data analysis and interpretation and reviewing and editing of the manuscript. All authors provided critical feedback and helped shape the research, analysis and manuscript.
Corresponding author
Ethics declarations
Competing interests
M.C. receives funding from the National Institute for Health and Care Research (NIHR), UKRI, NIHR Biomedical Research Centre, NIHR Surgical Reconstruction and Microbiology Research Centre, NIHR Applied Research Collaboration West Midlands, NIHR Birmingham-Oxford Blood and Transplant Research Unit in Precision Transplant and Cellular Therapeutics, UK SPINE, European Regional Development Fund—Demand Hub and Health Data Research UK at the University of Birmingham and University Hospitals Birmingham NHS Foundation Trust, Innovate UK (part of UKRI), Macmillan Cancer Support, UCB Pharma, GSK and Gilead. M.C. has received personal fees from Astellas, Aparito, CIS Oncology, Takeda, Merck, Daiichi Sankyo, Glaukos, GSK and the Patient-Centered Outcomes Research Institute outside the submitted work. S.K. is a consultant for Hardian Health and vice president of Conceivable Life Sciences. S.R.P., N.R., H.C.-L. and K.H. are employees of Google. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks Evangelos Evangelou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Lorenzo Righetto and Saheli Sadanand, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Supplementary information
Supplementary Information
Scoping survey questions.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Arora, A., Alderman, J.E., Palmer, J. et al. The value of standards for health datasets in artificial intelligence-based applications. Nat Med 29, 2929–2938 (2023). https://doi.org/10.1038/s41591-023-02608-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-023-02608-w
This article is cited by
-
A causal perspective on dataset bias in machine learning for medical imaging
Nature Machine Intelligence (2024)
-
Clinical applications of artificial intelligence in robotic surgery
Journal of Robotic Surgery (2024)