Abstract
B cells, which are critical for intestinal homeostasis, remain understudied in ulcerative colitis (UC). In this study, we recruited three cohorts of patients with UC (primary cohort, n = 145; validation cohort 1, n = 664; and validation cohort 2, n = 143) to comprehensively define the landscape of B cells during UC-associated intestinal inflammation. Using single-cell RNA sequencing, single-cell IgH gene sequencing and protein-level validation, we mapped the compositional, transcriptional and clonotypic landscape of mucosal and circulating B cells. We found major perturbations within the mucosal B cell compartment, including an expansion of naive B cells and IgG+ plasma cells with curtailed diversity and maturation. Furthermore, we isolated an auto-reactive plasma cell clone targeting integrin αvβ6 from inflamed UC intestines. We also identified a subset of intestinal CXCL13-expressing TFH-like T peripheral helper cells that were associated with the pathogenic B cell response. Finally, across all three cohorts, we confirmed that changes in intestinal humoral immunity are reflected in circulation by the expansion of gut-homing plasmablasts that correlates with disease activity and predicts disease complications. Our data demonstrate a highly dysregulated B cell response in UC and highlight a potential role of B cells in disease pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All requests for raw and analyzed data and materials will be promptly reviewed by the corresponding author and the study team. We have provided source data files for all the figures. Raw sequencing reads of scRNA-seq samples, as well as UMI tables, are available in the Gene Expression Omnibus under accession number GSE182270. Additional raw data are provided in Supplementary Tables 1–22. Source data are provided with this paper.
Code availability
The R code developed for clustering and analyses in this study is available at our GitHub page: https://github.com/effiken/Uzzan_et_al. Source data are provided with this paper.
References
Ungaro, R., Mehandru, S., Allen, P. B., Peyrin-Biroulet, L. & Colombel, J.-F. Ulcerative colitis. Lancet 389, 1756–1770 (2016).
Macpherson, A. J. & Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004).
Peterson, D. A., McNulty, N. P., Guruge, J. L. & Gordon, J. I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Bollinger, R. R. et al. Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Mol. Immunol. 43, 378–387 (2006).
Geissmann, F. et al. A subset of human dendritic cells expresses IgA Fc receptor (CD89), which mediates internalization and activation upon cross-linking by IgA complexes. J. Immunol. 166, 346–352 (2001).
Pilette, C., Detry, B., Guisset, A., Gabriels, J. & Sibille, Y. Induction of interleukin-10 expression through Fcα receptor in human monocytes and monocyte-derived dendritic cells: role of p38 MAPKinase. Immunol. Cell Biol. 88, 486–493 (2010).
Phalipon, A. & Corthésy, B. Novel functions of the polymeric Ig receptor: well beyond transport of immunoglobulins. Trends Immunol. 24, 55–58 (2003).
Donaldson, G. P. et al. Gut microbiota utilize immunoglobulin a for mucosal colonization. Science 360, 795–800 (2018).
McLoughlin, K., Schluter, J., Rakoff-Nahoum, S., Smith, A. L. & Foster, K. R. Host selection of microbiota via differential adhesion. Cell Host Microbe 19, 550–559 (2016).
Kabbert, J. et al. High microbiota reactivity of adult human intestinal IgA requires somatic mutations. J. Exp. Med. 217, e20200275 (2020).
Castro-Dopico, T. & Clatworthy, M. R. IgG and Fcγ receptors in intestinal immunity and inflammation. Front. Immunol. 10, 805 (2019).
Castro-Dopico, T. et al. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity 50, 1099–1114 (2019).
MacDermott, R. P. et al. Alterations in serum immunoglobulin G subclasses in patients with ulcerative colitis and Crohn’s disease. Gastroenterology 96, 764–768 (1989).
Ueno, H., Banchereau, J. & Vinuesa, C. G. Pathophysiology of T follicular helper cells in humans and mice. Nat. Immunol. 16, 142–152 (2015).
Xue, G. et al. Aberrant alteration of follicular T helper cells in ulcerative colitis patients and its correlations with interleukin-21 and B cell subsets. Medicine (Baltimore) 98, e14757 (2019).
Rao, D. A. et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017).
Martin, J. C. et al. Single-cell analysis of crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508 (2019).
Castro-Dopico, T., Colombel, J. F. & Mehandru, S. Targeting B cells for inflammatory bowel disease treatment: back to the future. Curr. Opin. Pharmacol. 55, 90–98 (2020).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 1–12 (2017). 2017 81.
Portugal, S. et al. Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. eLife 4, e07218 (2015).
Jenks, S. A. et al. Distinct effector B cells induced by unregulated Toll-like receptor 7 contribute to pathogenic responses in systemic Lupus erythematosus. Immunity 49, 725–739 (2018).
Dann, E., Henderson, N. C., Teichmann, S. A., Morgan, M. D. & Marioni, J. C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-01033-z (2021).
Kett, K. & Brandtzaeg, P. Local IgA subclass alterations in ulcerative colitis and Crohn’s disease of the colon. Gut 28, 1013–21 (1987).
Snapper, C. M. & Paul, W. E. Interferon-γ and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science 236, 944–947 (1987).
Toellner, K. M. et al. T helper 1 (Th1) and Th2 characteristics start to develop during T cell priming and are associated with an immediate ability to induce immunoglobulin class switching. J. Exp. Med. 187, 1193–1204 (1998).
Gene Set Enrichment Analysis. Molecular Signatures Database (MSigDB). https://www.gsea-msigdb.org/gsea/msigdb
Obeng-Adjei, N. et al. Malaria-induced interferon-γ drives the expansion of Tbethi atypical memory B cells. PLoS Pathog. 13, e1006576 (2017).
Landsverk, O. J. B. et al. Antibody-secreting plasma cells persist for decades in human intestine. J. Exp. Med. 214, 309–317 (2017).
Benckert, J. et al. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J. Clin. Invest. 121, 1946–1955 (2011).
Kuwada, T. et al. Identification of an anti-integrin αvβ6 autoantibody in patients with ulcerative colitis. Gastroenterology 160, 2383–2394 (2021).
Hovhannisyan, Z., Treatman, J., Littman, D. R. & Mayer, L. Characterization of interleukin-17-producing regulatory T cells in inflamed intestinal mucosa from patients with inflammatory bowel diseases. Gastroenterology 140, 957–965 (2011).
Habtezion, A., Nguyen, L. P., Hadeiba, H. & Butcher, E. C. Leukocyte trafficking to the small intestine and colon. Gastroenterology 150, 340–354 (2016).
Cole, K. E. et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J. Exp. Med. 187, 2009–2021 (1998).
Katewa, A. et al. Btk-specific inhibition blocks pathogenic plasma cell signatures and myeloid cell-associated damage in IFNα-driven lupus nephritis. JCI insight 2, e90111 (2017).
Owczarczyk, K. et al. A plasmablast biomarker for nonresponse to antibody therapy to CD20 in rheumatoid arthritis. Sci. Transl. Med. 3, 101ra92 (2011).
Brandtzaeg, P. et al. The B-cell system of human mucosae and exocrine glands. Immunol. Rev. 171, 45–87 (1999).
James, K. R. et al. Distinct microbial and immune niches of the human colon. Nat. Immunol. 21, 343–353 (2020).
Boland, B. S. et al. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci. Immunol. 5, eabb4432 (2020).
Baklien, K. & Brandtzaeg, P. Comparative mapping of the local distribution of immunoglobulin-containing cells in ulcerative colitis and Crohn’s disease of the colon. Clin. Exp. Immunol. 22, 197–209 (1975).
Macpherson, A., Khoo, U. Y., Forgacs, I., Philpott-Howard, J. & Bjarnason, I. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut 38, 365–375 (1996).
Uo, M. et al. Mucosal CXCR4+ IgG plasma cells contribute to the pathogenesis of human ulcerative colitis through FcγR-mediated CD14 macrophage activation. Gut 62, 1734–1744 (2013).
Van Den Bogaerde, J. et al. Gut mucosal response to food antigens in Crohn’s disease. Aliment. Pharmacol. Ther. 16, 1903–1915 (2002).
Kett, K., Rognum, T. O. & Brandtzaeg, P. Mucosal subclass distribution of immunoglobulin G-producing cells is different in ulcerative colitis and Crohn’s disease of the colon. Gastroenterology 93, 919–924 (1987).
Dunn-Walters, D. K., Isaacson, P. G. & Spencer, J. Sequence analysis of human IgVH genes indicates that ileal lamina propria plasma cells are derived from Peyer’s patches. Eur. J. Immunol. 27, 463–467 (1997).
Ekland, E. H., Forster, R., Lipp, M. & Cyster, J. G. Requirements for follicular exclusion and competitive elimination of autoantigen-binding B cells. J. Immunol. 172, 4700–4708 (2004).
Yurasov, S. & Nussenzweig, M. C. Regulation of autoreactive antibodies. Curr. Opin. Rheumatol. 19, 421–426 (2007).
Schoenborn, J. R. & Wilson, C. B. Regulation of interferon-γ during innate and adaptive immune responses. Adv. Immunol. 96, 41–101 (2007).
Hosomi, S. et al. Increased numbers of immature plasma cells in peripheral blood specifically overexpress chemokine receptor CXCR3 and CXCR4 in patients with ulcerative colitis. Clin. Exp. Immunol. 163, 215–224 (2011).
Wang, X. et al. The shifted balance between circulating follicular regulatory T cells and follicular helper T cells in patients with ulcerative colitis. Clin. Sci. 131, 2933–2945 (2017).
Long, Y. et al. The imbalance of circulating follicular helper T cells and follicular regulatory T cells is associated with disease activity in patients with ulcerative colitis. Front. Immunol. 11, 104 (2020).
Wang, Z. et al. Circulating follicular helper T cells in Crohn’s disease (CD) and CD-associated colorectal cancer. Tumor Biol. 35, 9355–9359 (2014).
Faubion, W. A. et al. EMerging BiomARKers in Inflammatory Bowel Disease (EMBARK) study identifies fecal calprotectin, serum MMP9, and serum IL-22 as a novel combination of biomarkers for Crohn’s disease activity: role of cross-sectional imaging. Am. J. Gastroenterol. 108, 1891–1900 (2013).
Amir, EadD., Guo, X. V., Mayovska, O. & Rahman, A. H. Average overlap frequency: a simple metric to evaluate staining quality and community identification in high dimensional mass cytometry experiments. J. Immunol. Methods 453, 20–29 (2018).
Van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytom. Part A 87, 636–645 (2015).
Maecker, H. T., McCoy, J. P. & Nussenblatt, R. Standardizing immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol. 12, 191–200 (2012).
Finak, G. et al. Standardizing flow cytometry immunophenotyping analysis from the Human ImmunoPhenotyping Consortium. Sci. Rep. 6, 20686 (2016).
Wardemann, H. & Kofer, J. Expression cloning of human B cell immunoglobulins. Methods Mol. Biol. 971, 93–111 (2013).
Alexander, K. L. et al. Human microbiota flagellins drive adaptive immune responses in Crohn’s disease. Gastroenterology 161, 522–535 (2021).
effiken/scDissector. https://github.com/effiken/scDissector
Baran, Y. et al. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 20, 206 (2019).
Jaitin, D. A. et al. Massively parallel single cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Paul, F. et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163, 1663–1677 (2015).
Mahnke, Y. D., Brodie, T. M., Sallusto, F., Roederer, M. & Lugli, E. The who’s who of T-cell differentiation: human memory T-cell subsets. Eur. J. Immunol. 43, 2797–2809 (2013).
Savas, P. et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 24, 986–993 (2018).
Zemmour, D. et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR article. Nat. Immunol. 19, 291–301 (2018).
Zheng, C. et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342–1356 (2017).
Hayatsu, N. et al. Analyses of a mutant Foxp3 allele reveal BATF as a critical transcription factor in the differentiation and accumulation of tissue regulatory T cells. Immunity 47, 268–283 (2017).
Förster, R., Davalos-Misslitz, A. C. & Rot, A. CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 8, 362–371 (2008).
Willinger, T. et al. Human naive CD8 T cells down-regulate expression of the WNT pathway transcription factors lymphoid enhancer binding factor 1 and transcription factor 7 (T cell factor-1) following antigen encounter in vitro and in vivo. J. Immunol. 176, 1439–1446 (2006).
Hombrink, P. et al. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 17, 1467–1478 (2016).
Kumar, B. V. et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017).
Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016).
Thome, J. J. C. et al. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat. Med. 22, 72–77 (2016).
Wong, M. T. et al. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity 45, 442–456 (2016).
Chattopadhyay, P. K., Yu, J. & Roederer, M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat. Med. 11, 1113–1117 (2005).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 14, 7 (2013).
Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010).
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Acknowledgements
We thank the patients who participated in the study. The authors would like to thank the Biorepository and Pathology Core at the Icahn School of Medicine at Mount Sinai for carrying out some of the immunostaining experiments. This work was supported by NIH/NIDDK grant R01 112296 (S.M.). Additional support was provided by a Rainin Foundation Synergy Grant to S.M., G.J.R. and J.F.C. A.C. was supported by P01 AI061093 and Ministerio de Ciencia, Innovación y Universidades grants RTI2018-093894-B-I00 and RTI2018-093894-B-I00. Other sources of support included K23KD111995 (R.C.U.), a Career Development Award from the Crohn’s and Colitis Foundation (R.C.U.), R01 AI119006 (G.D.V.), R01 AI157137 (G.D.V.), DK121009 (S.A.L.) and DK110352 (S.A.L.). M.U. was supported by an ECCO-IOBD fellowship, by a grant from the French National Society of Gastroenterology (Bourse Robert Tournut) and by the Fondation pour la Recherche Médicale (FDM 41552). J.C.M. was supported by ‘Prix pour les jeunes chercheurs’ de la Fondation Bettencourt-Schueller, the Philippe Foundation, NExT ‘Junior Talent’ and ANR JCJC (ANR-20-CE17-0009). E.K.G. was supported by a postdoctoral fellowship from the Swedish Research Council (2015-06486). Q.Z. is the recipient of a research fellowship from Takeda Pharmaceuticals. M.R.C. and T.D.C. were supported by a Medical Research Council New Investigator Research Grant (MR/N024907/1) and by a Wellcome Trust Investigator Award (220268/Z/20/Z). The sampling of the IBD cohort (Crohn’s disease and UC) was jointly designed as part of the research alliance between Janssen Biotech and the Icahn School of Medicine at Mount Sinai. Beyond this exception, no other funders had a role in analyses, design or interpretation.
Author information
Authors and Affiliations
Contributions
S.M., M.U. and J.M. drafted the manuscript. S.M. designed the study, supervised experimental data collection and coordinated integration of collaboration between all participating laboratories. All other authors contributed to experimental data and analyses, and, along with S.M., M.U. and J.M., they critically reviewed and edited the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.M. and J.F.C. have an unrestricted, investigator-initiated grant from Takeda Pharmaceuticals to examine novel homing mechanisms to the gastrointestinal tract. R.C.U. has served as an advisory board member or consultant for Eli Lilly, Janssen, Pfizer and Takeda. Mount Sinai coauthors (from Genetics and Genomics, Icahn Institute for Data Science and Genomic Technology, Human Immune Monitoring Center, Population Health Science and Policy, Division of Gastroenterology, Pediatric GI and Hepatology, Susan and Leonard Feinstein IBD Clinical Center at the Icahn School of Medicine at Mount Sinai) were partially funded as part of the research alliance between Janssen Biotech and the Icahn School of Medicine at Mount Sinai. J.P. and C.B. are employees of Janssen Research and Development, and J.R.F. is a former employee of Janssen Research and Development and is currently employed at Alnylam Pharmaceuticals. M.K., N.R., W.O. and J.H. are all employees of Genentech. M.D. is a consultant for Janssen, Abbvie, Arena, Bristol Meyers Squibb, Boehringer Ingelheim, Eli Lilly, Janssen, Pfizer, Prometheus Labs and Takeda. C.E. has received research support from Merck. R.U. has served as an advisory board member or consultant for AbbVie, Bristol Meyers Squibb, Janssen, Pfizer and Takeda and has received research support from AbbVie, Boehringer Ingelheim, Eli Lilly and Pfizer. J.F.C. reports receiving research grants from AbbVie, Janssen Pharmaceuticals and Takeda; receiving payment for lectures from AbbVie, Amgen, Allergan, Ferring Pharmaceuticals, Shire and Takeda; receiving consulting fees from AbbVie, Amgen, Arena Pharmaceuticals, Boehringer Ingelheim, Bristol Myers Squibb, Celgene Corporation, Eli Lilly, Ferring Pharmaceuticals, Galmed Research, GlaxoSmithKline, Geneva, Iterative Scopes, Janssen Pharmaceuticals, Kaleido Biosciences, Landos, Otsuka, Pfizer, Prometheus, Sanofi, Takeda and TiGenix; and holding stock options in Intestinal Biotech Development. B.L.C. receives the following financial support: advisory boards and consultant for Abbvie, Celgene/Bristol Myers Squibb, Eli Lilly, Pfizer, Sublimity Therapeutics, Takeda and TARGET RWE; CME companies: Cornerstones and Vindico; speaking fees: Abbvie; and educational grant: Pfizer. B.S. discloses research grants from Takeda, Pfizer, Theravance Biopharma R&D and Janssen; consulting fees from 4D Pharma, Abivax, Abbvie, Alimentiv, Allergan, Amgen, Arena Pharmaceuticals, AstraZeneca, Bacainn Therapeutics, Boehringer Ingelheim, Boston Pharmaceuticals, Bristol Myers Squibb, Calibr, Capella Bioscience, Celgene, Celltrion Healthcare, ClostraBio, Enthera, F. Hoffmann-La Roche, Ferring, Galapagos, Gilead, GlaxoSmithKline, GossamerBio, Immunic, Index Pharmaceuticals, Innovation Pharmaceuticals, Ironwood Pharmaceuticals, Janssen, Kaleido, Kallyope, Eli Lilly, MiroBio, Morphic Therapeutic, Oppilan Pharma, OSE Immunotherapeutics, Otsuka, Palatin Technologies, Pfizer, Progenity, Prometheus Biosciences, Prometheus Laboratories, Protagonist Therapeutics, Q32 Bio, Redhill Biopharma, Rheos Medicines, Salix Pharmaceuticals, Seres Therapeutics, Shire, Sienna Biopharmaceuticals, Sun Pharma, Surrozen, Takeda, Target PharmaSolutions, Teva Branded Pharmaceutical Products R&D, Thelium, Theravance Biopharma R&D, TLL Pharma, USWM Enterprises, Ventyx Biosciences, Viela Bio, Vivante Health and Vivelix Pharmaceuticals; and stock in Vivante Health and Ventyx Biosciences. All other authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks Jose Ordovas-Montanes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Saheli Sadanand was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Outline of the patients and samples included in the study.
HC: Healthy controls; aUC: active ulcerative colitis; qUC: quiescent ulcerative colitis; PBMC = peripheral blood mononuclear cells.
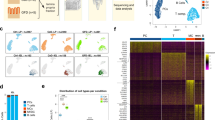
Extended Data Fig. 2 Lineage grouping within 7 major cellular compartments from single-cell RNA sequencing of colonic biopsies.
a, Grouping single-cell RNA sequencing cell clusters by similarity. Heatmap showing Pearson correlation coefficients between the log-averaged expression profiles of clusters. Ordering was determined by hierarchical clustering, which unbiasedly grouped the clusters by cellular lineage (color-coded bar). b, Heatmap showing color-coded down-sampled UMI counts of representative lineage marker genes in 100 randomly selected cells per cluster. The color code on the left indicates the lineage annotation of the different clusters as follows: red=T cells; orange=innate lymphoid cells; blue= plasma cells; yellow= B cells; green= mononuclear phagocytes; purple- mast cells; teal= stromal cells. c, Major cellular compartment subtypes frequencies within total lamina propria cells. The number of cells in each cellular subtype is divided by the total number of lamina propria cells and visualized in a stacked bar for each sample (HC and patients with UC).
Extended Data Fig. 3 Characterization of the 20 colonic clusters of B cells from single-cell RNA sequencing of colonic biopsies.
a, UMI counts in B lymphocyte and plasma cell clusters. box plots represent the third quartile (top of the box), median (center lines) and first quartile (bottom of the box) of measurements, and the whiskers represent 1.5 times the interquartile range from the top or bottom of the box. b, Heat-map showing color-coded down-sampled UMI counts of highly-variable genes between clusters of B cells (n = 7621 cells) and PC (n = 21425 cells). Clusters are demarcated by gray bars. Cells were down-sampled to 2000 UMIs/cell. Detailed cell counts per B and PB/PC cluster per sample are accessible in Supplementary Table 2. c, Correlation matrix of gene modules defined by gene-to-gene correlation analysis of single cells within the B lymphocyte clusters. d, Heat-map showing color-coded down-sampled UMI counts of gene programs identifying naïve, memory and germinal center-like B lymphocytes. Clusters are demarcated by gray bars. Cells were down-sampled to 2000 UMIs/cell. e,I FN signature scores on the indicated cell types. Supplementary Table 2 shows cell-counts per cluster, box plots represent the third quartile (top of the box), median (center lines) and first quartile (bottom of the box) of measurements, and the whiskers represent 1.5 times the interquartile range from the top or bottom of the box.
Extended Data Fig. 4 JH, VH family usage and clonal relationships.
a-d, IgH V and J gene family usage in short-lived colonic PC (a,c) and in circulating gut-homing PBs (b,d). Data are shown as individual data, mean and standard deviation; Mann- Whitney test, two tailed. e, Mean IgH amino acid CDR3 length per patient excluding JH6 + clones. Mann-Whitney test, two tailed, no significant difference. Panels a-e are based on 4 HC and 4 active UC patients f, Clonal relationships of all clones analyzed for each HC and UC patient. Numbers within circles stand for total number of clones.
Extended Data Fig. 5 Serological reactivity against microbiota antigens.
Thirty-two bacterial proteins and extracts (described in detail in Supplementary Table 8) were arrayed in triplicate on 16 pad nitrocellulose slides. After blocking, patient serum was added to the respective pads to determine reactivity against these antigens. a-b, Representation of increased IgG (a) and IgA (b) reactivity against microbial antigens 14-2 F, M18-1, COE1 and EcLW in patients with UC. c, IgG and IgA serological reactivity against 32 microbial antigens. Data are shown as individual data, mean and standard deviation, t-test, two-tailed. Details of the data from panel c are also provided in Supplementary Table 7.
Extended Data Fig. 6 Generation of a molecular signature of an IgG-activated or LPS colonic macrophage activation response.
a, Generation of a molecular signature of IgG-activated macrophages using transcriptomic analysis of murine colonic macrophages stimulated ex vivo with IgG immune complexes. The signature was restricted to human- and disease-relevant genes using a second dataset of IgG IC-stimulated human monocytes (GSE102728) and elastic-net regularized logistic regression on a training mucosal UC dataset (GSE38713), respectively. A second microbial gene signature using transcriptomic analysis of colonic macrophages from DSS-treated mice and refined on LPS-treated human monocytes was also generated. b, List of constituent genes for the immune complexes signature and the LPS signature. The right panel shows the number of exclusive and shared genes between the 2 signatures.
Extended Data Fig. 7 Proportion of “resident-like” and inflammatory macrophages and their respective molecular signature.
a, Proportion of “resident-like” macrophages and inflammatory macrophages among total colonic macrophages represented in individual samples of healthy controls UC and patients. b, Heat-map showing color-coded down-sampled UMI counts of gene programs identifying LPS signature (red), immune complexes (IC) signature (green) and resident macrophage genes signatures (black).
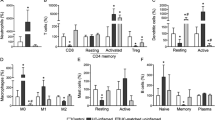
Extended Data Fig. 8 Characterization of T cell clusters from single-cell RNA sequencing of colonic biopsies.
a, Heat-map showing the relative expression of 5 transcriptional scores (rows) (see Methods) in single T cells (n = 11281 cells) (columns). Cell clusters are demarcated by gray bars. Detailed cell counts per T cluster per sample are accessible in Supplementary Table 2. b, Euclidean distance for lineage normalized cell type frequencies was compared within healthy controls (HC), (HC-HC, blue), patients with UC (UC-UC, red) and between HC and patients with UC (HC-UC, grey) for T cells, Wilcoxon rank-sum test. c, Relative expression of genes identifying T cell subsets. Heatmap is showing log-2 transformed expression per subset divided by the mean expression across all subsets.
Extended Data Fig. 9 Representative histological examination of a surgical specimen from a UC patient before immunofluorescent staining.
Macroscopic view of colon after resection. Black arrow delineates disease margin, which runs strikingly anti-parallel to the length of the colon. b, H&E section showing disease margin (yellow) line with proximal region on the left and margin progressing toward disease distally on the right. Note the increased number of small follicles on the diseased side. c, Normal isolated lymphoid follicle in the colon. d, Proximal colon from UC patient with delimited disease in the region that appears macroscopically healthy, with abnormal fibrotic accumulation beneath the epithelial lining. e, Macroscopically normal proximal specimen from a UC patient that contains diffuse inflammatory infiltrate despite a healthy macroscopic appearance. For panels b-e, white bars = 200 μm.
Supplementary information
Source data
Source Data
Relevant source data for figures
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Uzzan, M., Martin, J.C., Mesin, L. et al. Ulcerative colitis is characterized by a plasmablast-skewed humoral response associated with disease activity. Nat Med 28, 766–779 (2022). https://doi.org/10.1038/s41591-022-01680-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-022-01680-y
This article is cited by
-
Deciphering the different phases of preclinical inflammatory bowel disease
Nature Reviews Gastroenterology & Hepatology (2024)
-
Anti-integrin αvβ6 autoantibodies are a potential biomarker for ulcerative colitis-like immune checkpoint inhibitor-induced colitis
British Journal of Cancer (2024)
-
Identification of FCN1 as a novel macrophage infiltration-associated biomarker for diagnosis of pediatric inflammatory bowel diseases
Journal of Translational Medicine (2023)
-
A precise molecular subtyping of ulcerative colitis reveals the immune heterogeneity and predicts clinical drug responses
Journal of Translational Medicine (2023)
-
Single-cell meta-analysis of inflammatory bowel disease with scIBD
Nature Computational Science (2023)