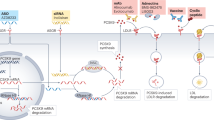
Abstract
Compelling evidence supports a causal role for lipoprotein(a) (Lp(a)) in cardiovascular disease. No pharmacotherapies directly targeting Lp(a) are currently available for clinical use. Here we report the discovery and development of olpasiran, a first-in-class, synthetic, double-stranded, N-acetylgalactosamine-conjugated small interfering RNA (siRNA) designed to directly inhibit LPA messenger RNA translation in hepatocytes and potently reduce plasma Lp(a) concentration. Olpasiran reduced Lp(a) concentrations in transgenic mice and cynomolgus monkeys in a dose-responsive manner, achieving up to over 80% reduction from baseline for 5–8 weeks after administration of a single dose. In a phase 1 dose-escalation trial of olpasiran (ClinicalTrials.gov: NCT03626662), the primary outcome was safety and tolerability, and the secondary outcomes were the change in Lp(a) concentrations and olpasiran pharmacokinetic parameters. Participants tolerated single doses of olpasiran well and experienced a 71–97% reduction in Lp(a) concentration with effects persisting for several months after administration of doses of 9 mg or higher. Serum concentrations of olpasiran increased approximately dose proportionally. Collectively, these results validate the approach of using hepatocyte-targeted siRNA to potently lower Lp(a) in individuals with elevated plasma Lp(a) concentration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
GENCODE data are available from the following gzipped file: http://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_24/gencode.v24.annotation.gtf.gz. Other preclinical data that support the findings reported in this paper are available from the corresponding authors upon reasonable request.
Regarding the clinical trial portions of the paper, qualified researchers may request data from Amgen clinical studies. Complete details are available at https://wwwext.amgen.com/science/clinical-trials/clinical-data-transparencypractices/clinical-trial-data-sharing-request/.
References
Schmidt, K., Noureen, A., Kronenberg, F. & Utermann, G. Structure, function, and genetics of lipoprotein (a). J. Lipid Res. 57, 1339–1359 (2016).
Kronenberg, F. & Utermann, G. Lipoprotein(a): resurrected by genetics. J. Intern. Med. 273, 6–30 (2013).
Eckardstein, A. V. Lipoprotein(a). Eur. Heart J. 38, 1530–1532 (2017).
Langsted, A., Kamstrup, P. R. & Nordestgaard, B. G. High lipoprotein(a) and high risk of mortality. Eur. Heart J. 40, 2760–2770 (2019).
Clarke, R. et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 361, 2518–2528 (2009).
Kamstrup, P. R., Tybjaerg-Hansen, A., Steffensen, R. & Nordestgaard, B. G. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA 301, 2331–2339 (2009).
Nordestgaard, B. G. et al. Lipoprotein(a) as a cardiovascular risk factor: current status. Eur. Heart J. 31, 2844–2853 (2010).
Helgadottir, A. et al. Apolipoprotein(a) genetic sequence variants associated with systemic atherosclerosis and coronary atherosclerotic burden but not with venous thromboembolism. J. Am. Coll. Cardiol. 60, 722–729 (2012).
Thanassoulis, G. et al. Associations of long-term and early adult atherosclerosis risk factors with aortic and mitral valve calcium. J. Am. Coll. Cardiol. 55, 2491–2498 (2010).
Kamstrup, P. R., Tybjaerg-Hansen, A. & Nordestgaard, B. G. Elevated lipoprotein(a) and risk of aortic valve stenosis in the general population. J. Am. Coll. Cardiol. 63, 470–477 (2014).
Kral, B. G. et al. Relation of plasma lipoprotein(a) to subclinical coronary plaque volumes, three-vessel and left main coronary disease, and severe coronary stenoses in apparently healthy African-Americans with a family history of early-onset coronary artery disease. Am. J. Cardiol. 118, 656–661 (2016).
Thanassoulis, G. Lipoprotein (a) in calcific aortic valve disease: from genomics to novel drug target for aortic stenosis. J. Lipid Res. 57, 917–924 (2016).
Tsimikas, S. A test in context: lipoprotein(a): diagnosis, prognosis, controversies, and emerging therapies. J. Am. Coll. Cardiol. 69, 692–711 (2017).
Gudbjartsson, D. F. et al. Lipoprotein(a) concentration and risks of cardiovascular disease and diabetes. J. Am. Coll. Cardiol. 74, 2982–2994 (2019).
Patel, A. P. et al. Lp(a) (lipoprotein[a]) concentrations and incident atherosclerotic cardiovascular disease: new insights from a large national biobank. Arterioscler. Thromb. Vasc. Biol. 41, 465–474 (2021).
Yahya, R. et al. Statin treatment increases lipoprotein(a) levels in subjects with low molecular weight apolipoprotein(a) phenotype. Atherosclerosis 289, 201–205 (2019).
Artemeva, N. V. et al. Lowering of lipoprotein(a) level under niacin treatment is dependent on apolipoprotein(a) phenotype. Atheroscler. Suppl. 18, 53–58 (2015).
Baum, S. J. et al. Effect of evolocumab on lipoprotein apheresis requirement and lipid levels: results of the randomized, controlled, open-label DE LAVAL study. J. Clin. Lipidol. 13, 901–909 (2019).
Nugent, A., Gray, J., Gorby, L. & Moriarty, P. Lipoprotein apheresis: first FDA indicated treatment for elevated lipoprotein (a). J. Clin. Cardiol. 1, 16–21 (2019).
Shen, X. & Corey, D. R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 46, 1584–1600 (2018).
Watts, J. K. & Corey, D. R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 226, 365–379 (2012).
Roberts, T. C., Langer, R. & Wood, M. J. A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 19, 673–694 (2020).
Varvel, S., McConnell, J. P. & Tsimikas, S. Prevalence of elevated Lp(a) mass levels and patient thresholds in 532 359 patients in the United States. Arterioscler. Thromb. Vasc. Biol. 36, 2239–2245 (2016).
Marcovina, S. M. et al. Differences in Lp[a] concentrations and apo[a] polymorphs between black and white Americans. J. Lipid Res. 37, 2569–2585 (1996).
Burgess, S. et al. Association of LPA variants with risk of coronary disease and the implications for lipoprotein(a)-lowering therapies: a Mendelian randomization analysis. JAMA Cardiol. 3, 619–627 (2018).
Lamina, C. & Kronenberg, F. Lp(a)-GWAS-Consortium. Estimation of the required lipoprotein(a)-lowering therapeutic effect size for reduction in coronary heart disease outcomes: a Mendelian randomization analysis. JAMA Cardiol. 4, 575–579 (2019).
Madsen, C. M., Kamstrup, P. R., Langsted, A., Varbo, A. & Nordestgaard, B. G. Lipoprotein(a)-lowering by 50 mg/dL (105 nmol/L) may be needed to reduce cardiovascular disease 20% in secondary prevention: a population-based study. Arterioscler. Thromb. Vasc. Biol. 40, 255–266 (2020).
Tsimikas, S. et al. Lipoprotein(a) reduction in persons with cardiovascular disease. N. Engl. J. Med. 382, 244–255 (2020).
Chi, X., Gatti, P. & Papoian, T. Safety of antisense oligonucleotide and siRNA-based therapeutics. Drug Discov. Today 22, 823–833 (2017).
Judge, D. P. et al. Phase 3 multicenter study of revusiran in patients with hereditary transthyretin-mediated (hATTR) amyloidosis with cardiomyopathy (ENDEAVOUR). Cardiovasc. Drugs Ther. 34, 357–370 (2020).
Springer, A. D. & Dowdy, S. F. GalNAc-siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 28, 109–118 (2018).
Kenski, D. M. et al. siRNA-optimized modifications for enhanced in vivo activity. Mol. Ther. Nucleic Acids 1, e5 (2012).
Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019).
Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162 (2019).
Kozomara, A. & Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2014).
Kozomara, A. & Griffiths-Jones, S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 (2011).
Griffiths-Jones, S., Saini, H. K., van Dongen, S. & Enright, A. J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158 (2008).
Griffiths-Jones, S. The microRNA registry. Nucleic Acids Res. 32, D109–D111 (2004).
Zhang, G., Budker, V. & Wolff, J. A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 10, 1735–1737 (1999).
Wooddell, C. I., Reppen, T., Wolff, J. A. & Herweijer, H. Sustained liver-specific transgene expression from the albumin promoter in mice following hydrodynamic plasmid DNA delivery. J. Gene Med. 10, 551–563 (2008).
Dati, F. et al. First WHO/IFCC international reference reagent for lipoprotein(a) for immunoassay—Lp(a) SRM 2B. Clin. Chem. Lab. Med. 42, 670–676 (2004).
Marcovina, S. M., Albers, J. J., Gabel, B., Koschinsky, M. L. & Gaur, V. P. Effect of the number of apolipoprotein(a) kringle 4 domains on immunochemical measurements of lipoprotein(a). Clin. Chem. 41, 246–255 (1995).
Marcovina, S. M. et al. Use of a reference material proposed by the International Federation of Clinical Chemistry and Laboratory Medicine to evaluate analytical methods for the determination of plasma lipoprotein(a). Clin. Chem. 46, 1956–1967 (2000).
Yeang, C., Witztum, J. L. & Tsimikas, S. ‘LDL-C’ = LDL-C + Lp(a) − C: implications of achieved ultra-low LDL-C levels in the proprotein convertase subtilisin/kexin type 9 era of potent LDL-C lowering. Curr. Opin. Lipidol. 26, 169–178 (2015).
Willeit, P. et al. Low-density lipoprotein cholesterol corrected for lipoprotein(a) cholesterol, risk thresholds, and cardiovascular events. J. Am. Heart Assoc. 9, e016318 (2020).
Langlois, M. R. et al. Quantifying atherogenic lipoproteins for lipid-lowering strategies: consensus-based recommendations from EAS and EFLM. Clin. Chem. Lab. Med. 58, 496–517 (2020).
Acknowledgements
Amgen, Inc. and Arrowhead Pharmaceuticals, Inc. funded this work. J. Murray, H. Hamilton, M. Stolina, D. Dwyer and P. Denis supported the in vitro screening and preclinical data. J. Hegge performed some of the preclinical work (mouse and monkey) and supported data preparation for the manuscript. P. Yamaguchi assisted in drafting the preclinical section of the manuscript. J. Wang and the Amgen Clinical Biomarker and Diagnostics group supported Lp(a) assay evaluation and selection. N. Shah and Z. Atter provided statistical support for the clinical study. W. Krall of Wanda Krall Medical Communications, funded by Amgen, and E. Stoltzfus of Amgen provided editorial support.
Author information
Authors and Affiliations
Contributions
M.J.K., P.M.M., S.J.B., J.N., M.H.-I., H.S.W. and G.F.W. enrolled participants in the clinical study and supervised the clinical components of the trial (clinical review, safety and dose monitoring). M.J.K., P.M.M., S.J.B. and G.F.W. additionally interpreted results and critically revised the manuscript. H.W. and M.E.-D. performed statistical planning and analysis of the clinical study, interpreted the results and critically revised the manuscript. W.S. oversaw the clinical pharmacokinetic and pharmacodynamic data analyses, interpreted the results and critically revised the manuscript. J.H. was involved in the oversight of the clinical study (for example, eligibility, protocol amendments and safety monitoring), study data analysis, interpreting results and revision and drafting of the manuscript. T.V. was involved in safety monitoring, study data analysis, interpreting results and revision of the manuscript. S.M.H. was involved in conceptualization and drafting and revision of the manuscript and interpreting results. H.K. was involved in study data analysis, interpreting results and revision and drafting of the manuscript. B.M.R. oversaw the cynomolgus monkey study and data analysis and interpreted results. M.F. and S.M. were involved in the design and interpretation of the preclinical studies and prepared the preclinical portion of the manuscript. M.F. directed additional safety and manufacturability assessments to select the clinical candidate olpasiran. T.P. designed the siRNA molecules for the in vitro and in vivo screening. O.H. conducted the bioinformatics analysis. All authors reviewed the manuscript, approved the final version and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
M.J.K. is an employee of Jacksonville Center for Clinical Research, an organization that has received research grant funds from Amgen and multiple other manufacturers of lipid-lowering agents. M.J.K. has no direct personal financial relationship with Amgen, the manufacturer of olpasiran, and is an unpaid member of the Northeast Florida AHA Board. P.M.M. reports speaker and consulting fees from Amgen and Regeneron; consulting/advisory fees and grant research support from Esperion; grant research support from Ionis, Aegerion and the FH Foundation; speaker fees from Amarin; consulting fees from Stage II Innovations/Renew and Kaneka; advisor fees from Novartis; and fees for genetic testing kits from GB Life Sciences. S.J.B. reports Scientific Advisory Board/Consultant/Speaker for Amgen; and Scientific Advisory Board/Consultant for Sanofi Regeneron, Akcea and Novartis. J.N. and M.H.-I. have nothing to disclose. H.S.W. reports research funding from Amgen and Novo Nordisk; research funding and scientific advisory fees from Novartis; and speaker fees from Esperion. S.M. and T.P. are employees of Arrowhead Pharmaceuticals and have received Arrowhead stock options. M.F., H.K., T.V., W.S., H.W., M.E.D., B.M.R., O.H. and J.H. are employees of Amgen and own Amgen stock. S.M.H. is an officer, executive and shareholder in Amgen; a scientific founder of and shareholder in Tenaya Therapeutics; and serves on the Scientific Advisory Board of the German Centre for Cardiovascular Research. G.F.W. has received honoraria for lectures or advisory boards and research grants from Sanofi, Regeneron, Amgen, Arrowhead, Novartis and Kowa.
Reviewer recognition statement Nature Medicine thanks Kosh Ray, Kathy Wolski and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Editor recognition statement Michael Basson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1
Phase 1 study schema.
Supplementary information
Supplementary Information
Supplementary Tables 1, 2 and 4
Supplementary Table 3
Transcript sequences were derived from the GENCODE version 24 (BASIC; https://www.gencodegenes.org/) gene model and the human GRCh38 reference genome, downloaded from the OmicSoft Studio software platform (Qiagen). Olpasiran guide and passenger sequences were aligned to the transcript sequences using a simple custom gapless aligner, employing a brute force approach in which the siRNA query sequence was exhaustively scanned as a sliding window across all transcript sequences. miRNA sequences were downloaded from miRBase (version 21; https://www.mirbase.org), and seed sequences (positions 2–7) were checked for human miRNA seed sequences matching the six-nucleotide seed region of the olpasiran guide sequences.
Rights and permissions
About this article
Cite this article
Koren, M.J., Moriarty, P.M., Baum, S.J. et al. Preclinical development and phase 1 trial of a novel siRNA targeting lipoprotein(a). Nat Med 28, 96–103 (2022). https://doi.org/10.1038/s41591-021-01634-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-021-01634-w
This article is cited by
-
An LC–MS-based designated comparison method with similar performance to the Lp(a) reference measurement procedure to guide molar Lp(a) standardization
Clinical Proteomics (2024)
-
Targeting Lipoprotein(a): Can RNA Therapeutics Provide the Next Step in the Prevention of Cardiovascular Disease?
Cardiology and Therapy (2024)
-
Lipoprotein(a) in patients with breast cancer after chemotherapy: exploring potential strategies for cardioprotection
Lipids in Health and Disease (2023)
-
Calcific aortic valve disease: mechanisms, prevention and treatment
Nature Reviews Cardiology (2023)
-
From target discovery to clinical drug development with human genetics
Nature (2023)