Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.1.7 and B.1.351 variants were first identified in the United Kingdom and South Africa, respectively, and have since spread to many countries. These variants harboring diverse mutations in the gene encoding the spike protein raise important concerns about their immune evasion potential. Here, we isolated infectious B.1.1.7 and B.1.351 strains from acutely infected individuals. We examined sensitivity of the two variants to SARS-CoV-2 antibodies present in sera and nasal swabs from individuals infected with previously circulating strains or who were recently vaccinated, in comparison with a D614G reference virus. We utilized a new rapid neutralization assay, based on reporter cells that become positive for GFP after overnight infection. Sera from 58 convalescent individuals collected up to 9 months after symptoms, similarly neutralized B.1.1.7 and D614G. In contrast, after 9 months, convalescent sera had a mean sixfold reduction in neutralizing titers, and 40% of the samples lacked any activity against B.1.351. Sera from 19 individuals vaccinated twice with Pfizer Cominarty, longitudinally tested up to 6 weeks after vaccination, were similarly potent against B.1.1.7 but less efficacious against B.1.351, when compared to D614G. Neutralizing titers increased after the second vaccine dose, but remained 14-fold lower against B.1.351. In contrast, sera from convalescent or vaccinated individuals similarly bound the three spike proteins in a flow cytometry-based serological assay. Neutralizing antibodies were rarely detected in nasal swabs from vaccinees. Thus, faster-spreading SARS-CoV-2 variants acquired a partial resistance to neutralizing antibodies generated by natural infection or vaccination, which was most frequently detected in individuals with low antibody levels. Our results indicate that B1.351, but not B.1.1.7, may increase the risk of infection in immunized individuals.
Similar content being viewed by others
Main
SARS-CoV-2 variants have rapidly emerged in humans and supplanted ancestral strains1,2,3,4,5. Their proposed increased rates of interindividual transmission conferred a replication advantage at the population level. One of the first identified variants includes the D614G mutation in the gene encoding the spike (S) protein, which enhances viral infectivity and shifts S protein conformation toward an angiotensin-converting enzyme 2 (ACE2)-binding fusion-competent state, without significantly modifying sensitivity to antibody neutralization1,6,7,8. More recently, novel variants have appeared in multiple countries, with combinations of mutations and deletions in the receptor-binding domain (RBD) and N-terminal domain of S protein, as well as in other proteins. The B.1.1.7 variant emerged in the United Kingdom, the B.1.351 variant (also termed 501Y.V2) in South Africa and the P.1 and P.2 lineages in Brazil2,3,5,9,10,11,12. Although distinct, the variants share common characteristics, including known escape mutations that were previously identified under antibody pressure selection in vitro2,3,13,14,15,16,17. Some of the mutations or deletions were also identified in immunocompromised individuals with prolonged infectious viral shedding and treated with convalescent plasma or S-protein monoclonal antibodies3,17,18,19, indicating that antibody-escape mutations are selected in vivo. The sensitivity to antibody neutralization varies with the viral variant. B.1.1.7 seems to be more sensitive to neutralization than B.1.351. The RBD mutation N501Y, which increases affinity to ACE2 and is present in B.1.1.7 and B.1.351 (ref. 20), does not impair on its own post-vaccine serum neutralization21. It has been suggested that the other mutations in B.1.1.7 do not result in immune evasion of linear epitopes22. Mutations in the B.1.351 and P.1 strains, including E484K and K417N/T, are of high concern, since they partly compromise neutralization generated by previous infection or vaccination23,24,25 or may increase inherent viral fitness. The Pfizer Cominarty (also termed BNT162b2) vaccine-elicited human sera neutralizes SARS-CoV-2 lineage B.1.1.7 pseudovirus, with slightly reduced titers in some vaccinees, when compared to the Wuhan reference strain26,27. The Moderna mRNA-1273 vaccine also induces neutralizing antibodies to SARS-CoV-2 variants with, however, a five- to ten-fold reduction in efficacy against the B.1.351 S protein, when compared to pseudovirus bearing the D614G mutation27,28.
Neutralization efficacy has so far mostly been assessed using vesicular stomatis virus (VSV)-derived or lentivirus-derived pseudovirus assays, or with infectious SARS-CoV-2 carrying point mutations in S protein. A recent report using infectious B.1.351 virus showed that plasma samples from six convalescent donors were strongly attenuated against this strain, with half-maximal inhibitory concentration (IC50) values 6- to 200-fold higher than ancestral virus29. It is thus of utmost importance to use authentic variant strains in addition to pseudovirus particles, since mutations outside of the S protein may impact inherent viral fitness and/or sensitivity to antibodies. Here, we compared the sensitivity of three authentic SARS-CoV-2 strains, the preexisting D614G virus and the B.1.1.7 and B.1.351 variants, to antibody neutralization.
Results
Isolation and characterization of SARS-CoV-2 B.1.1.7 and B.1.351 variants
We isolated the two variants from nasal swabs of individuals with infection diagnosed by quantitative PCR with reverse transcription (RT–qPCR) and sequence-diagnosed infection. The viruses were amplified by one to two passages on Vero cells. Sequences of the outgrown viruses confirmed the identity of B.1.1.7 and B.1.351 variants. Viral stocks were titrated using S-Fuse reporter cells30. These cells are derived from a human immortalized line, termed U2OS-ACE2+ cells. They carry the GFP-split complementation system, in which two cells separately produce half of the reporter protein, producing GFP only upon fusion. Following infection, the cells produce S protein at their surface and fuse with neighboring cells, generating a GFP signal as soon as 6 h after infection30 (Fig. 1a). The number of GFP+ cells correlated with the viral inoculum (Fig. 1b). Neutralizing SARS-CoV-2 monoclonal antibodies (mAbs) targeting the RBD can be classified into four main categories31,32. Using the S-Fuse assay, we tested the sensitivity of the three viral strains to two RBD antibodies, mAb102 and mAb48, derived from convalescent individuals. The antibodies belong to the first category and act by blocking binding of the ‘up’ conformation of RBD to ACE2 (ref. 32). MAb102 efficiently and similarly neutralized the three viral strains, with an IC50 of ≃0.01 µg ml−1 (Fig. 1c,d). MAb48 neutralized D614G (IC50 of 0.1 µg ml−1) but was inactive against B.1.1.7 and B.1.351 variants (Fig. 1c,d). These results confirm that the two variants can selectively display reduced sensitivity to certain antibodies. Of note, the activity of the monoclonal antibodies was similar at different viral inocula, within a range of 50 to 200 GFP+ syncytia per well (Fig. 1e), indicating that potential variations in the number of infected S-Fuse cells do not impact the calculation of IC50. Thus, for further studies, we selected a multiplicity of infection leading to about 150 syncytia per well for each viral strain.
a, Principle of the S-Fuse reporter assay. S-Fuse cells, a mix of U20S-ACE2-GFP1–10 and U20S-ACE2-GFP11, were cocultured at a 1:1 ratio and infected with SARS-CoV-2. Cells fused upon infection and generated a GFP signal. Infection was quantified by measuring the number of GFP+ syncytia at 18 h. b, Linearity of the assay with D614G (gray), B.1.1.7 (blue) and B.1.351 (green) strains. S-Fuse cells were exposed to serial dilutions of viral stocks and infection was quantified by high-content imaging. c, Representative images (from five independent experiments) of S-Fuse cells infected with D614G, B.1.1.7 and B1.351 strains in the presence or absence of two SARS-CoV-2 monoclonal antibodies (mAb48 and mAb102). Scale bar, 400 µm. d, Dose-response analysis of the neutralization by mAb102 and mAb48 on the three viral strains. e, Neutralization of SARS-CoV-2 (B.1.1.7) by mAb102 at three viral inocula. Results are shown as the mean ± standard deviation (s.d.) from four independent experiments.
Sensitivity of D614G, B.1.1.7 and B.1.351 variants to sera from convalescent individuals
We assessed the neutralization ability of sera from convalescent individuals. We selected samples from 28 donors in a longitudinal cohort of individuals with SARS-CoV-2 from the French city of Orléans (Supplementary Table 1). All individuals were diagnosed with SARS-CoV-2 infection by RT–qPCR or serology and including those who had critical, severe and mild-to-moderate coronavirus disease 2019 (COVID-19). They were not vaccinated. A total of 25 individuals were sampled twice: first, at a median of 89 d (range 77–105 d) post onset of symptoms (POS; 3-month samples (M3)), and second, at a median of 179 d (range 168–197 d) POS (6-month samples (M6). We incubated serially diluted sera with D614G, B.1.1.7 or B.1.351 strains, added the mixture to S-Fuse cells, and scored the GFP+ cells after overnight infection. We then calculated the median effective dose (ED50) for each combination of serum and virus. A representative example with the same donor is depicted in Fig. 2a, and the results with all donors appear in Fig. 2b. The D614G and B.1.1.7 strains were similarly sensitive to sera. At M3, the ED50 values were 1.5 × 103 and 1 × 103 for D614G and B.1.1.7 variants, respectively, with huge variations between individuals (from 102 to 2 × 104), and the values did not strongly decline at M6. As expected, individuals with critical disease displayed higher neutralizing activities than those with severe or mild-to-moderate symptoms (Extended Data Fig. 1). Again, there was no significant difference between the two viral strains in each category of symptoms. For B1.351, the neutralization titers were significantly decreased by five and ten-fold at the two time points, when compared with D614G and B.1.1.7 strains (Fig. 2b and Extended Data Fig. 1).
a, Examples of neutralization curves with sera from two donors (D18 and D59). The first donor, from the Orléans cohort, was sequentially sampled at months 3 (M3) and 6 (M6; left and middle, respectively). The second donor, from the Strasbourg cohort, was sampled at month 9 (M9; right). Results are shown as the mean ± s.d. from three independent experiments. b, ED50 of neutralization of the three viral isolates. Sera from the Orléans cohort were sequentially sampled at M3 (n = 28) and M6 (n = 25; left and middle, respectively), and 30 sera from the Strasbourg cohort were sampled at M9 (right). Data are the mean from two to four independent experiments. The dotted line indicates the limit of detection (ED50 = 30). Two-sided Friedman test with Dunn’s test for multiple comparisons was performed between each viral strain at the different time points; **P < 0.01, ***P < 0.001, ****P < 0.0001. M3: D614G versus B.1.351, P = 0.0012; B.1.1.7 versus B.1.351, P = 0.0012. M6: D614G versus B.1.351, P = 0.0005; B.1.1.7 versus B.1.351, P < 0.0001. M9: D614G versus B.1.351, P < 0.0001; B.1.1.7 versus B.1.351, P < 0.0001. c, Each individual was arbitrarily defined as a ‘neutralizer’ (blue) if neutralizing activity was detected at the first serum dilution (1:30) or ‘non-neutralizer’ (gray) if no activity was detected. The numbers indicate the percentage of neutralizers.
We confirmed these results in sera from 30 individuals from another cohort of RT–qPCR-confirmed unvaccinated health-care workers from Strasbourg University Hospital who experienced mild disease33,34. The samples were collected at a later time point (M9), with a median of 233 d (range 206–258 d) POS (Supplementary Table 1). Overall, the neutralization activity was lower (one representative example is shown in Fig. 2a and the results of all donors are available in Fig. 2b). There was no significant difference in neutralization between D614G and B.1.1.7 strains, with a similar ED50 of 2 × 102. In sharp contrast, the neutralizing activity against B.1.351 was particularly low at this late time point, with a median ED50 of 50, representing a fourfold decrease when compared to D614G (Fig. 2b).
We then arbitrarily classified the individuals as neutralizers (with neutralizing antibodies detectable at the first serum dilution of 1:30) and non-neutralizers, for the three viral strains and the two cohorts (Fig. 2c). Most individuals neutralized the three strains at M3. The fraction of neutralizers started to decline at M6, a phenomenon which was more marked for B.1.351. The fraction of neutralizers was higher in the second cohort, with 93% of individuals neutralizing either D614G or B.1.1.7 strains at M9. In contrast, only 63% of individuals neutralized the B.1.351 strain (Fig. 2c).
Antibody binding to cells expressing D614G, B.1.1.7 and B.1.351 S proteins
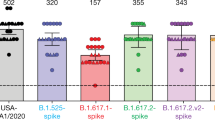
We next examined the binding capacity of monoclonal antibodies and sera from the convalescent individuals described above to S proteins from different lineages. To this aim, we adapted the flow cytometry-based S-Flow assay, which we previously established to measure the levels of antibodies binding to cells expressing the Wuhan S protein35. We transiently transfected human epithelial HEK293T (referred to as 293T) cells with plasmids expressing the D614G, B.1.1.7 and B.1.351 S proteins. Similar surface levels of the viral proteins were detected with mAb10, a non-neutralizing pan-coronavirus antibody targeting a conserved epitope in the S2 domain and isolated from a convalescent individual (C. Planchais et al., unpublished data; Fig. 3a and Supplementary Fig. 1). Moreover, the cells fused when cocultivated with ACE2-expressing cells, indicating that the three proteins were functional (data not shown). mAb48 efficiently bound to D614G S proteins, slightly less potently to B.1.1.7, and lost all binding activity to B.1.351. Therefore, the escape of mAb48 neutralization by the two variants is due to mutations decreasing or abrogating antibody binding to its target. mAb102 bound similarly to the three S proteins, in accordance with its cross-reactive neutralizing activity (Fig. 3a,b). The N501Y mutation enhances RBD affinity to ACE2, when tested with recombinant proteins and yeast surface display20. Using flow cytometry, we assessed by the binding of a labeled soluble ACE2 protein to cells expressing the different S proteins. We observed a dramatic increase in binding of soluble ACE2 to B.1.1.7 and to a lesser extent to B.1.351, which both carry the N501Y mutation, when compared to D614G (Fig. 3a,b).
a, Binding of monoclonal antibodies or soluble ACE2. HEK293T cells were transiently transfected with plasmids expressing the D614G, B.1.1.7 and B.1.351 S proteins. After 24 h, cells were stained with SARS-CoV-2 antibody mAb10 (a pan-coronavirus antibody), mAb48, mAb102 or soluble ACE2 (ACE2-biotin at 10 μg ml−1 revealed with fluorescent streptavidin) and analyzed by flow cytometry. One representative example of binding is shown. b, Titration binding curves of mAb48, mAb102 and ACE2 to the three S proteins. Data are the mean ± s.d. of three independent experiments. c, Binding of the panel of 83 sera from 58 convalescent individuals. Sera were tested at a 1:300 dilution. Data are the mean of two independent experiments. Two-sided Friedman test with Dunn’s test for multiple comparisons was performed between each viral strain at the different time points, *P < 0.05. M6: B.1.1.7 versus B.1.351, P = 0.037.
We next tested our panel of sera from convalescent individuals against the different S proteins. Overall, the sera bound similarly to all three S proteins, even though B.1.351 displayed a slight reduction (statistically significant at M6) in the mean fluorescence intensity (MFI) of binding (Fig. 3c). We observed a global and slight decrease in MFI at M9, compared to samples from M3 and M6 (Fig. 3c). Altogether, these results indicate that S proteins from B.1.351, and to a higher extent B.1.1.7, display increased affinity for ACE2 while escaping binding to some monoclonal antibodies and either to a lesser degree or not at all to polyclonal sera.
Sensitivity of D614G, B.1.1.7 and B.1.351 variants to sera from vaccine recipients
We next asked whether vaccine-elicited antibodies inhibit infection by the different variants. In France, vaccination started in January 2020 with the Pfizer Cominarty vaccine. We thus selected 19 vaccine recipients from a cohort of vaccinated health-care workers established in Orléans. The characteristics of vaccinees are depicted in Supplementary Table 2. Sera and nasal swabs were sampled at week 2 (17 individuals), week 3 (18 individuals), week 4 (11 individuals; corresponding to week 1 after the second dose), week 5 (16 individuals; corresponding to week 2 after the second dose) and week 6 (15 individuals; corresponding to week 3 after the second dose), with a median of 13, 19, 28, 34 and 41 d after the first dose, respectively. This allowed us to assess the early humoral response to vaccination. We first analyzed 16 of 19 vaccinees that were not previously infected with SARS-CoV-2, as assessed by the absence of preexisting S-protein and anti-nucleoprotein (N) antibodies. A representative example of the evolution of the neutralizing response in one vaccine recipient at the four time points is depicted in Fig. 4a. In this instance, the serum only neutralized D614G at week 2, whereas B.1.1.7 started to be neutralized at week 3, although less efficiently than D614G. B.1.1.7 and D614G strains were similarly neutralized at week 4. The anti-B.1.351 response was negative up to week 3, and became detectable at week 4, at a level lower than the two other viruses.
a, Examples of neutralization curves with sera from one donor (D9) at week 2 (W2), W3, W4 (W1 after second dose) and W6 (W3 after second dose) after vaccination. Results are shown as the mean ± s.d. from three independent experiments. b, Neutralization ED50 values of the three viral isolates. Sera from 10 to 16 vaccine recipients were sampled at W2 (n = 15), W3 (n = 16) and W1 and W3 after the second dose (n = 10 and 15, respectively). Data are the mean from two to four independent experiments. The dotted line indicates an ED50 of 30. Two-sided Friedman test with Dunn’s test for multiple comparisons was performed between each viral strain at the different time points; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. W2: D614G versus B.1.1.7, P = 0.0105; D614G versus B.1.351, P = 0.008. W3: D614G versus B.1.351, P = 0.0002. W4: D614G versus B.1.351, P = 0.011; B.1.1.7 versus B.1.351, P = 0.0052. W6: D614G versus B.1.1.7, P = 0.0324; D614G versus B.1.351, P = 0.0324; B.1.1.7 versus B.1.351, P < 0.0001. c, Evolution overtime of neutralizing antibody titers in 14 vaccine recipients. The arrows indicate the first and second doses of vaccines. d, Each individual was arbitrarily defined as a ‘neutralizer’ (blue) if neutralizing activity was detected at a 1:30 serum dilution or ‘non-neutralizer’ (gray) if no activity was detected. The numbers indicate the percentage of neutralizers.
The ED50 results from sera of the 16 vaccine recipients at the four time points are presented in Fig. 4b, whereas the evolution of their neutralization titers over the 6 weeks after vaccination appears in Fig. 4c. We also arbitrarily classified individuals as neutralizers (with neutralizing antibodies detectable at a serum dilution of 1:30) and non-neutralizers, for the three viral strains (Fig. 4d). Two weeks after vaccination, antibodies neutralizing the D614G strain were detected in sera from 5 of 15 recipients (33%; with an arbitrary threshold of ED50 > 30 for neutralization positivity; Fig. 4b,d). The titers were relatively modest at this early time point (ED50 of 30). These low titers were less efficient against B.1.1.7, with 2 of 15 neutralizers (13%), and were inactive against B.1.351 (Fig. 4b,c). At week 3, the neutralizing activity increased against D614G and B.1.1.7. There was, however, a threefold reduction in the neutralization titers against B.1.1.7 (Fig. 4b), whereas B.1.351 remained insensitive. At the last time point (week 3 after the second dose), titers increased in most of the recipients and were similar between D614G and B.1.1.7. Titers remained 14-fold and 53-fold lower against B.1.351, when compared to D614G and B.1.117, respectively (Fig. 4b,c). At week 3, 63%, 38% and 0% of the samples neutralized D614G, B.1.1.7 and B.1.351 strains, respectively (Fig. 4d). At week 6, 92% of the samples neutralized D614G and B.1.1.7, whereas 77% neutralized B.1.351 with a low titer (Fig. 4d). The S-Flow assay demonstrated the presence of antibodies binding to the three S proteins at the different sampling times (Extended Data Fig. 2). Therefore, the Pfizer Cominarty vaccine generated a neutralizing response that efficiently targeted D614G and B.1.1.7 with a delay in the appearance of neutralizing antibodies to B.1.1.7 and B.1.351 relative to D614G. The titers remained lower against B.1.351, even in responders. The vaccine displays a protective efficacy against COVID-19 as soon as 2 weeks after the first dose36. Our results suggest that the low neutralizing titers (ED50 of 50–100) in the sera may correspond to the observed protection against severe disease.
Sensitivity of SARS-CoV-2 B.1.1.7 and B.1.351 variants to nasal swabs from vaccine recipients
Little is known about the levels and function of vaccine-elicited antibodies in mucosal samples. We thus measured the neutralizing activity and the levels of antibodies in nasal swabs from the series of vaccine recipients (Extended Data Fig. 2). We did not detect any antiviral effect in these samples at all time points (up to 6 weeks after vaccination; Extended Data Fig. 2a,b). We also analyzed three additional vaccine recipients who were seropositive for anti-N at the time of vaccination, indicative of a previous infection. Two recipients were diagnosed with SARS-CoV-2 by PCR in March and April 2020 and experienced a mild disease. The third recipient did not report any previous signs reminiscent of COVID-19. In these three individuals, the serum neutralizing titers were strikingly high at week 2 after the first dose (ED50 of about 104; Extended Data Fig. 2c) and remained similarly high at week 5 (data not shown). Two of the three vaccine recipients who were previously seropositive for SARS-CoV-2 displayed a low neutralizing activity in their nasal swabs, when measured at week 2 after the first dose (Extended Data Fig. 2d). Their nasal swabs neutralized D614G and B.1.117 strains similarly but were inactive against B.1.351. As a control, we analyzed ten pre-pandemic nasopharyngeal swab specimens (collected by the National Reference Center in 2018 for the purpose of influenza surveillance) for the presence of neutralizing SARS-CoV-2 antibodies. The samples were all negative starting at a 1:4 dilution, ruling out a confounding nonspecific effect of a substance that may be present in the swabs (Supplementary Fig. 2). The levels of S-protein antibodies and their neutralization activity in the sera and nasal swabs of vaccine recipients are summarized in the heat map in Extended Data Fig. 2f. In the swabs, we detected SARS-CoV-2 S-protein-binding antibodies in 7 of 13 available samples at week 3, and 14 of 15 samples at week 4 (Extended Data Fig. 2f). The three previously seropositive vaccine recipients had the highest binding antibody levels in the nasal swab. Unfortunately, we did not have access to their nasal swabs before vaccination, precluding determination of whether previous infection, vaccination or, more likely, the combination of both, generated a neutralizing response in these individuals. Of note, the sera of the two previously infected and vaccinated individuals also contained neutralizing antibodies (Extended Data Fig. 2f) Altogether, our results suggest that vaccinees probably do not elicit an early humoral response detectable at mucosal surfaces. They strengthen the hypothesis that some vaccines may not protect against viral acquisition and infection of the oral–nasal region, but may prevent severe disease associated with viral dissemination in the lower respiratory tract. Our results are in line with those obtained in nonhuman primates, in which vaccinated and then challenged animals display detectable viral loads in nasal swabs but not in lower airways37.
Discussion
Here, we analyzed the cross-reactivity of the humoral response to preexisting SARS-CoV-2 viruses and recently emerging variants in sera from long-term convalescent individuals and recent vaccine recipients. We report that immune sera had slightly reduced but largely preserved activity against B.1.1.7 when compared to the reference D614G strain. The B.1.351 variant is potentially more problematic as it is less sensitive or even insensitive to sera from immunized individuals. The E484K mutation, which can mediate antibody escape of B.1.351 and P.1 variants, has been recently detected in the B.1.1.7 lineage in the United Kingdom, and thus represents a major potential threat for previously immune populations. Here, we used authentic clinical viral isolates, rather than pseudovirus, providing a relevant way to assess inherent viral fitness and the potential impact of additional mutations outside of S protein on sensitivity to neutralizing antibodies. On a technical note, the combination of S-Fuse and S-Flow assays, two methods of analysis of viral infectivity, neutralization or antibody levels, allowed a rapid assessment of the properties of emerging variants. The workflow established here can be easily adapted to any novel viral strain. Potential limitations of our study are the relatively low number of individuals analyzed and the short time frame of analysis after vaccination. Furthermore, we have not investigated the impact of preexisting cellular immunity, which may be more cross-reactive against variants than the humoral response. Future work with more vaccine recipients that have or haven’t been previously infected, and longer surveillance periods, will help to characterize the role of local and systemic humoral responses in vaccine efficacy. Our study also highlights the importance of the second dose of the Pfizer Cominarty vaccine, which was associated with a strong increase of neutralizing antibody titers and a widening of strain cross-reactive antibody responses. In conclusion, our results demonstrate that suboptimal or declining antibody responses are associated with a loss of cross-reactivity against novel emerging viral strains.
Methods
Orléans cohort of convalescent and vaccinated individuals
Since 27 August 2020, a prospective, monocentric, longitudinal, interventional cohort clinical study enrolling 170 SARS-CoV-2-infected individuals with different disease severities, and 30 noninfected healthy controls is ongoing, aiming to describe the persistence of specific and neutralizing antibodies over a 24-month period. This study was approved by the Ile-de-France IV ethical committee. At enrollment, written informed consent was collected and participants completed a questionnaire covering sociodemographic characteristics, virological findings (SARS-CoV-2 RT–qPCR results, including date of testing), clinical data (date of symptom onset, type of symptoms, hospitalization) and data related to anti-SARS-CoV-2 vaccination if applicable (brand product, date of first and second vaccination). Serological status of participants was assessed every 3 months. Those who underwent anti-SARS-CoV-2 vaccination had weekly blood and nasal sampling after the first dose of vaccine for a 2-month period (NCT04750720). For the present study, we selected 28 convalescent and 19 vaccinated participants. Nasal swabs were collected in the two cavities and preserved in 3 ml of transport M4RT buffer (Remel Microtask, Thermo Fisher). Study participants did not receive any compensation. Our research complied with all relevant ethical regulations.
Strasbourg cohort of convalescent individuals
Since 17 April 2020, a prospective, interventional, monocentric, longitudinal, cohort clinical study enrolling 308 RT–qPCR-diagnosed SARS-CoV-2-infected hospital staff from Strasbourg University Hospital is ongoing (NCT04441684). At enrollment, written informed consent was collected and participants completed a questionnaire covering sociodemographic characteristics, virological findings (SARS-CoV-2 RT–qPCR results including date of testing) and clinical data (date of symptom onset, type of symptoms, hospitalization). This study was approved by Institutional Review Board of Strasbourg University Hospital. The serological status of the participants has been described at M3 and M6 POS33,34. Laboratory identification of SARS-CoV-2 was performed at least 10 d before inclusion by RT–qPCR testing on nasopharyngeal swab specimens according to current guidelines (Institut Pasteur; WHO technical guidance). The assay targets two regions of the viral RNA-dependent RNA polymerase (RdRp) gene with a threshold of detection of ten copies per reaction. For the present study, we randomly selected 30 individuals collected at M9. Study participants did not receive any compensation. None of the participants were vaccinated at the time of blood draws.
Pre-pandemic nasal swabs
Samples were collected as part of approved ongoing influenza virus surveillance conducted by the National Reference Center for Respiratory Viruses at Institut Pasteur. The laboratory investigations were carried out in accordance with the General Data Protection Regulation (EU Regulation 2016/679 and Directive 95/46/EC) and the French data protection law (Law 78–17 on 6 January 1978 and Décret 2019–536 on 29 May 2019), which does not require a review by an ethics committee for the secondary use of samples collected for health-care purposes. In such case, the secondary use for research is authorized if the individuals have been informed of such secondary use (article L.1211-2 of the French Public Health Code).
S-Fuse neutralization assay
U2OS-ACE2 GFP1–10 or GFP 11 cells, also termed S-Fuse cells, become GFP+ cells when they are productively infected with SARS-CoV-2 (ref 30). Cells tested negative for mycoplasma. Cells were mixed (at a 1:1 ratio) and plated at 8 × 103 cells per well in a μClear 96-well plate (Greiner Bio-One). The indicated SARS-CoV-2 strains were incubated with monoclonal antibodies, sera or nasal swabs at the indicated concentrations or dilutions for 15 min at room temperature and added to S-Fuse cells. The nasal swabs and sera were heat inactivated 30 min at 56 °C before use. Then, 18 h later, cells were fixed with 2% paraformaldehyde, washed and stained with Hoechst (1:1,000 dilution; Invitrogen). Images were acquired with an Opera Phenix high-content confocal microscope (PerkinElmer). The GFP area and the number of nuclei were quantified using the Harmony software (PerkinElmer). The percentage of neutralization was calculated using the number of syncytia as the value with the following formula: 100 × (1 − (value with serum − value in ‘noninfected’)/(value in ‘no serum’ − value in ‘noninfected’)). Neutralizing activity of each sera was expressed as the ED50. ED50 values (in µg ml−1 for monoclonal antibodies and in dilution values for sera and nasal swabs) were calculated using a reconstructed curve with the percentage of neutralization at the different indicated concentrations. Of note, we previously reported a correlation between neutralization titers obtained with the S-Fuse reporter assay and a pseudovirus neutralization assay38.
Virus strains
The reference D614G strain (hCoV-19/France/GE1973/2020) was supplied by the National Reference Centre for Respiratory Viruses hosted by Institut Pasteur and headed by S.v.d.W. This viral strain was supplied through the European Virus Archive goes Global (EVAg) platform, a project that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement number 653316. The variant strains were isolated from nasal swabs on Vero cells and amplified by one or two passages on Vero cells. The B.1.1.7 strain originated from an individual in Tours (France) who returned from the United Kingdom. The B.1.351 strain (CNR 202100078) originated from an individual in Créteil (France). Both individuals provided informed consent for the use of their biological materials. Titration of viral stocks was performed on Vero E6 cells, with a limiting dilution technique allowing a calculation of the 50% tissue culture infectious dose, or on S-Fuse cells. Viruses were sequenced directly on nasal swabs and after one or two passages on Vero cells.
Spike-protein expression plasmids
A codon-optimized version of the SARS-Cov-2 spike gene from Wuhan reference strain (GenBank: QHD43416.1), was transferred into the phCMV backbone (GenBank: AJ318514), by replacing the VSV-G gene. The B.1.1.7 (Δ69-70, ΔY144, N501Y, A570D, D614G, P681H, T716I, S982A and D1118H) and B.1.351 (L18F, D80A, D215G, Δ242-244, K417N, E484K, N501Y, D614G and A701V) spike mutations were added in silico into the codon-optimized Wuhan reference strain and were ordered as synthetic genes (GeneArt, Thermo Fisher Scientific) and transferred into the phCMV backbone. The D614G S-protein was generated by introducing the corresponding mutation in the Wuhan reference strain using Q5 Site-Directed Mutagenesis Kit (NEB).
Commercial serological assays
The serum samples from the Strasbourg cohort were first tested at Hôpital de Strasbourg using two commercial assays: (1) a CE-marked LFA assay for detection of IgM and IgG against the SARS-CoV-2 RBD of the S protein developed by Biosynex (COVID-19 BSS IgG/IgM), which has a specificity of 99% and a sensitivity of 96% for samples >22 d POS; and (2) a CE-marked ELISA for detection of IgG against the full-length recombinant N protein from Epitope Diagnostics (EDITM Novel coronavirus COVID-19 IgG), which has a specificity of 96% and a sensitivity of 81% after 28 d POS. The vaccine recipients were tested for anti-S antibodies at Hôpital Européen Georges Pompidou with the following assays: SARS-CoV-2 IgG (Abbott) targeting SARS-CoV-2 N protein were performed on an Architect i2000SR analyzer (Abbott). Index value threshold for positivity was set to 1.4 as recommended. Beckman Coulter Access SARS-CoV-2 IgG assays targeting the RBD were performed on a UniCel DxI 800 Access Immunoassay System (Beckman Coulter). Index value threshold for positivity was set to 1 as recommended. Qualitative results and index values were used in the analysis for both assays.
S-Flow assay
The S-Flow assay was performed as described previously33,35. Briefly, 293T cells were acquired from the American Type Culture Collection (CRL‐3216) and tested negative for mycoplasma. The 293T cells were transfected with the indicated S-protein expression plasmids or a control plasmid using Lipofectamine 2000 (Life technologies). One day after, transfected cells were detached using PBS-EDTA and transferred into U-bottom 96-well plates (50,000 cells per well). Cells were incubated at 4 °C for 30 min with sera (1:300 dilution) or nasal swabs (1:50 dilution) in PBS containing 0.5% BSA and 2 mM EDTA, washed with PBS and stained using anti-IgG AF647 (1:600 dilution; Thermo Fisher). Cells were washed with PBS and fixed for 10 min using 4% paraformaldehyde. Data were acquired on an Attune Nxt instrument (Life Technologies). Stainings were also performed on control (293T-empty) cells. The specificity and sensitivity of the S-Flow serological assay were originally assessed with the Wuhan S protein using 253 pre-pandemic samples and 377 RT–qPCR-confirmed SARS-CoV-2 samples. The sensitivity is 99.2% with a 95% confidence interval of 97.69–99.78% and the specificity is 100% (98.5–100%)33,35. Results were analyzed with FlowJo 10.7.1 (Becton Dickinson). After testing 253 pre-pandemic samples, the positivity of a sample was defined as a specific binding above 40%. The specific binding was calculated as follows: 100 × (percentage binding 293T S protein − percentage binding 293T-empty)/(100 − percentage binding 293T-empty).
Antibodies and ACE2 ectodomain
Human SARS-CoV-2 monoclonal antibodies were cloned from S-specific blood memory B cells of convalescent individuals with COVID-19 (C. Planchais et al., unpublished data). Monoclonal antibodies mAb48 and mAb102 recognize the RBD, and mAb10 binds to a conserved region of the S2 domain of the S protein. Human anti-S IgG monoclonal antibodies and His-tagged recombinant ACE2 ectodomain (amino acids 19–615) cloned into pcDNA3.1 vector were produced by transient transfection of Freestyle 293-F cells and purified by affinity chromatography as previously described39. Purified ACE2 protein was biotinylated using the EZ-Link Sulfo-NHS-Biotin kit (Thermo Fisher Scientific).
Statistical analysis
Flow cytometry data were analyzed with FlowJo v10 (TriStar). Calculations were performed using Excel 365 (Microsoft). Figures were drawn using Prism 9 (GraphPad Software). Statistical analysis was conducted using GraphPad Prism 9. Statistical significance between different groups was calculated using the tests indicated in each figure legend. No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. Source data are provided with this paper. Viral sequences are available upon request and were deposited at GISAID (https://www.gisaid.org/) under the following numbers: hCoV-19/France/GE1973/2020 (D614G): EPI_ISL_414631; B.1.1.7: EPI_ISL_735391; B.1.1.351: EPI_ISL_964916.
References
Yurkovetskiy, L. et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183, 739–751 (2020).
Kemp, S. et al. Recurrent emergence and transmission of a SARS-CoV-2 spike deletion ΔH69/V70. Preprint at BioRxiv https://doi.org/10.1101/2020.12.14.422555 (2020).
Kemp, S. A. et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature https://doi.org/10.1038/s41586-021-03291-y (2021).
Oude Munnink, B. B. et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371, 172–177 (2021).
Tegally, H. et al. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature https://doi.org/10.1038/s41586-021-03402-9 (2021).
Korber, B. et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827 (2020).
Plante, J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature https://doi.org/10.1038/s41586-020-2895-3 (2020).
Weissman, D. et al. D614G spike mutation increases SARS-CoV-2 susceptibility to neutralization. Cell Host Microbe 29, 23–31 (2021).
Rambaut, A. et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (2020).
Buss, L. F. et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 371, 288–292 (2021).
Faria, N. R. et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 (2021).
Sabino, E. C. et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet https://doi.org/10.1016/S0140-6736(21)00183-5 (2021).
Weisblum, Y. et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 9, e61312 (2020).
Andreano, E. et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. Preprint at BioRxiv https://doi.org/10.1101/2020.12.28.424451 (2020).
Rees-Spear, C. et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. https://doi.org/10.1016/j.celrep.2021.108890 (2021).
Liu, Z. et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29, 477–488.e4 (2021).
Starr, T. N. et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science https://doi.org/10.1126/science.abf9302 (2021).
Avanzato, V. A. et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell https://doi.org/10.1016/j.cell.2020.10.049 (2020).
McCarthy, K. R. et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 371, 1139–1142 (2021).
Zahradník, J. et al. SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor. Preprint at BioRxiv https://doi.org/10.1101/2021.01.06.425392 (2021).
Xie, X. et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. https://doi.org/10.1038/s41591-021-01270-4 (2021).
Haynes, W. A., Kamath, K., Lucas, C., Shon, J. & Iwasaki, A. Impact of B.1.1.7 variant mutations on antibody recognition of linear SARS-CoV-2 epitopes. Preprint at medRxiv https://doi.org/10.1101/2021.01.06.20248960 (2021).
Wibmer, C. K. et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. https://doi.org/10.1038/s41591-021-01285-x (2021).
Greaney, A. J. et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476.e6 (2021).
Thomson, E. C. et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell https://doi.org/10.1016/j.cell.2021.01.037 (2021).
Muik, A. et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science https://doi.org/10.1126/science.abg6105 (2021).
Wang, P. et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature https://doi.org/10.1038/s41586-021-03398-2 (2021).
Wu, K. et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. Preprint at BioRxiv https://doi.org/10.1101/2021.01.25.427948 (2021).
Cele, S. et al. Escape of SARS-CoV-2 501Y.V2 variants from neutralization by convalescent plasma. Preprint at medRxiv https://doi.org/10.1101/2021.01.26.21250224 (2021).
Buchrieser, J. et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 39, e106267 (2020).
Liu, L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020).
Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020).
Fafi-Kremer, S. et al. Serologic responses to SARS-CoV-2 infection among hospital staff with mild disease in eastern France. EBioMedicine https://doi.org/10.1016/j.ebiom.2020.102915 (2020).
Grzelak, L. et al. Sex differences in the evolution of neutralizing antibodies to SARS-CoV-2. J. Infect. Dis. https://doi.org/10.1093/infdis/jiab127 (2021).
Grzelak, L. et al. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.abc3103 (2020).
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. https://doi.org/10.1056/nejmoa2034577 (2020).
Dagotto, G., Yu, J. & Barouch, D. H. Approaches and challenges in SARS-CoV-2 vaccine development. Cell Host Microbe 28, 364–370 (2020).
Sterlin, D. et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.abd2223 (2020).
Lorin, V. & Mouquet, H. Efficient generation of human IgA monoclonal antibodies. J. Immunol. Methods 422, 102–110 (2015).
Acknowledgements
We thank F. Rey and L. Chakrabarti (Institut Pasteur) for critical reading of the manuscript. We thank the patients who participated in this study; members of the Virus and Immunity Unit for discussions and help; N. Aulner and the UtechS Photonic BioImaging (UPBI) core facility (Institut Pasteur), a member of the France BioImaging network, for image acquisition and analysis; and M. Backovic (Institut Pasteur) for the ACE2 construct. We thank the DRCI, CIC, Médecine du travail and Pôle de Biologie teams (CHU de Strasbourg) for the management of the Strasbourg cohort and serology testing. Work in the laboratory of O.S. is funded by Institut Pasteur, Urgence COVID-19 Fundraising Campaign of Institut Pasteur, ANRS, the Vaccine Research Institute (ANR-10-LABX-77), Labex IBEID (ANR-10-LABX-62-IBEID), ANR/FRM Flash COVID PROTEO-SARS-CoV-2 and IDISCOVR. Work in UPBI is funded by grant ANR-10-INSB-04-01 and Région Ile-de-France program DIM1-Health. D.P. is supported by the Vaccine Research Institute. L.G. is supported by the French Ministry of Higher Education, Research and Innovation. The laboratory of H.M. is funded by the Institut Pasteur, the Milieu Intérieur Program (ANR-10-LABX-69-01), the INSERM, REACTing and EU (RECOVER) grants. The laboratory of S.v.d.W. is funded by Institut Pasteur, CNRS, Université de Paris, Santé publique France, Labex IBEID (ANR-10-LABX-62-IBEID), REACTing, EU grant RECOVER. The laboratory of S.F.-K. is funded by Strasbourg University Hospital (SeroCoV-HUS; PRI 7782), Programme Hospitalier de Recherche Clinique (PHRC N 2017-HUS no. 6997), the Agence Nationale de la Recherche (ANR-18-CE17-0028), Laboratoire d’Excellence TRANSPLANTEX (ANR-11-LABX-0070_TRANSPLANTEX) and Institut National de la Santé et de la Recherche Médicale (UMR_S 1109). Work in CHR Orléans is funded by COREVIH Centre Val de Loire. The funders of this study had no role in study design, data collection, analysis and interpretation or writing of the article.
Author information
Authors and Affiliations
Contributions
Experimental strategy, design and experiments: D.P., T.B., L.G., F.G.-B., I.S., F.P., C.P., J.B., M.M.R., E.B., S.v.d.W., T.P. and O.S.; vital materials: M.A., F.D., S.B., V.E., M.M., M.G., J.D.S., H.P., D.V., A.S., E.S.L., S.F.-K., K.S., H.M., L.H., S.v.d.W and T.P.; manuscript writing: D.P., T.B. and O.S.; manuscript editing: L.D., J.D., M.M.R., H.P., D.V., S.F.-K., K.S., H.M., L.H., S.v.d.W. and T.P.
Corresponding author
Ethics declarations
Competing interests
L.G., I.S., T.B., J.B., F.G.-B. and O.S. are coinventors on provisional patent no. US 63/020,063 entitled ‘S-Flow: a FACS-based assay for serological analysis of SARS-CoV-2 infection’ submitted by Institut Pasteur.
Additional information
Peer review information Nature Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Alison Farrell was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sensitivity of SARS-CoV-2 variants to sera from different convalescent individuals.
ED50 of neutralization of the D614G, B.1.1.7 and B.1.351. 25 sera from the Orléans Cohort were sequentially sampled at month 3 (M3) and M6 (a and b, respectively) and analyzed as described in Fig. 2 legend. Results are shown as mean from two to four independent experiments. The individuals are classified according to disease severity. The bar indicates the median of values within each column. Two-sided Friedman test with Dunn’s multiple comparison was performed between each viral strain at the different time points, *: p-value<0.05. M6 (Severe): D614G versus B.1.351 p = 0.0417; B.1.1.7 versus B.1.351 p = 0.011. M6 (Mild-to-Moderate): B.1.1.7 versus B.1.351 p = 0.0315.
Extended Data Fig. 2 Sensitivity of SARS-CoV-2 D614G, B.1.1.7 and B.1.351 variants to nasal swabs from early vaccine recipients.
a, Example of neutralization curves with nasal swab from one donor at week 3 (W3) post vaccination. No neutralization was detected. Similar results were obtained at the different times of sampling, up to W6 post-vaccination b, ED50 of neutralization of the three viral isolates, with nasal swabs from 16 vaccine recipients sampled at W2, W3 post first dose and then W1 and W3 post-second dose. The dotted line indicates the limit of detection (ED50 = 4). No neutralization was detected. Data are mean from two to four independent experiments. c,d, Analysis of the three COVID-19 vaccinated patients. Neutralizing antibody titers are shown in the serum (dotted line: ED50 = 30) and nasal swab (dotted line: ED50 = 4), at week 2 post vaccination e, Example of neutralization curves with nasal swab from one such COVID-19 vaccine recipient at week 2 post vaccination. Results are shown as mean ± SD from three independent experiments. f, Heat map summarizing the levels of anti-Spike antibodies and their neutralizing activity in the sera and nasal swabs of the 19 vaccinated individuals. Vac #17, 18 and 19 were previously infected by SARS-CoV-2. White boxes denote an absence of activity. n.d. = not determined.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Figures and statistical raw data.
Source Data Fig. 2
Figures and statistical raw data.
Source Data Fig. 3
Figures and statistical raw data.
Source Data Fig. 4
Figures and statistical raw data.
Source Data Extended Data Fig. 1
Figures and statistical raw data.
Source Data Extended Data Fig. 2
Figures and statistical raw data.
Rights and permissions
About this article
Cite this article
Planas, D., Bruel, T., Grzelak, L. et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 27, 917–924 (2021). https://doi.org/10.1038/s41591-021-01318-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-021-01318-5
This article is cited by
-
Molecular characterization of SARS-CoV-2 Omicron clade and clinical presentation in children
Scientific Reports (2024)
-
Data-driven recombination detection in viral genomes
Nature Communications (2024)
-
Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion
Nature Communications (2024)
-
Characterization of intrinsic and effective fitness changes caused by temporarily fixed mutations in the SARS-CoV-2 spike E484 epitope and identification of an epistatic precondition for the evolution of E484A in variant Omicron
Virology Journal (2023)
-
Antibody induction and immune response in nasal cavity by third dose of SARS-CoV-2 mRNA vaccination
Virology Journal (2023)