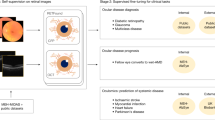
Abstract
Progression to exudative ‘wet’ age-related macular degeneration (exAMD) is a major cause of visual deterioration. In patients diagnosed with exAMD in one eye, we introduce an artificial intelligence (AI) system to predict progression to exAMD in the second eye. By combining models based on three-dimensional (3D) optical coherence tomography images and corresponding automatic tissue maps, our system predicts conversion to exAMD within a clinically actionable 6-month time window, achieving a per-volumetric-scan sensitivity of 80% at 55% specificity, and 34% sensitivity at 90% specificity. This level of performance corresponds to true positives in 78% and 41% of individual eyes, and false positives in 56% and 17% of individual eyes at the high sensitivity and high specificity points, respectively. Moreover, we show that automatic tissue segmentation can identify anatomical changes before conversion and high-risk subgroups. This AI system overcomes substantial interobserver variability in expert predictions, performing better than five out of six experts, and demonstrates the potential of using AI to predict disease progression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The clinical data used for the training, validation and test sets were collected at Moorfields Eye Hospital NHS Foundation Trust and transferred to DeepMind in a de-identified format. Data were used with both local and national permissions. They are not publicly available and restrictions apply to their use. The data, or a test subset, may be available from Moorfields Eye Hospital NHS Foundation Trust subject to local and national ethical approvals. Moorfields Eye Hospital NHS Foundation Trust intends to make the raw data shared with DeepMind openly available to researchers as part of the Ryan Initiative for Macular Research (http://rimr.doheny.org/).
Code availability
We made use of several open-source libraries to conduct our experiments, namely the machine learning framework TensorFlow (https://github.com/tensorflow/tensorflow) along with the TensorFlow library Sonnet (https://github.com/deepmind/sonnet), which provides implementations of individual model components53. For image augmentation we used the multidimension image augmentation library previously open sourced by DeepMind (https://github.com/deepmind/multidim-image-augmentation). The model architecture source code is available from (https://github.com/google-health/imaging-research). Other aspects of the experimental system made use of proprietary libraries and we are unable to publicly release this code. We detail the experiments and implementation details in Methods and in the Supplementary figures to allow for independent replication.
References
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
De Fauw, J. et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 24, 1342–1350 (2018).
Ardila, D. et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 25, 954–961 (2019).
Poplin, R. et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2, 158–164 (2018).
Tomašev, N. et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 572, 116–119 (2019).
Wong, W. L. et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob. Health 2, e106–e116 (2014).
Owen, C. G. et al. The estimated prevalence and incidence of late stage age related macular degeneration in the UK. Br. J. Ophthalmol. 96, 752–756 (2012).
Rein, D. B. et al. Forecasting age-related macular degeneration through the year 2050: the potential impact of new treatments. Arch. Ophthalmol. 127, 533–540 (2009).
Rudnicka, A. R. et al. Incidence of late-stage age-related macular degeneration in American whites: systematic review and meta-analysis. Am. J. Ophthalmol. 160, 85–93 (2015).
Lim, J. H. et al. Delay to treatment and visual outcomes in patients treated with anti-vascular endothelial growth factor for age-related macular degeneration. Am. J. Ophthalmol. 153, 678–686 (2012).
Bek, T. & Klug, S. E. Incidence and risk factors for neovascular age-related macular degeneration in the fellow eye. Graefes Arch. Clin. Exp. Ophthalmol. 256, 2061–2068 (2018).
Zarranz-Ventura, J. et al. The neovascular age-related macular degeneration database: report 2: incidence, management, and visual outcomes of second treated eyes. Ophthalmology 121, 1966–1975 (2014).
Fasler, K. et al. The Moorfields AMD Database Report 2 – Fellow Eye Involvement with Neovascular Age-related Macular Degeneration. Preprint at bioRxiv https://doi.org/10.1101/615252 (2019).
Maguire, M. G. et al. Incidence of choroidal neovascularization in the fellow eye in the comparison of age-related macular degeneration treatments trials. Ophthalmology 120, 2035–2041 (2013).
Amoaku, W. et al. Action on AMD. Optimising patient management: act now to ensure current and continual delivery of best possible patient care. Eye 26, S2–S21 (2012).
Chew, E. Y., Lindblad, A. S. & Clemons, T. Summary results and recommendations from the Age-Related Eye Disease Study. Arch. Ophthalmol. 127, 1678 (2009).
Cohen, S. Y. et al. Prevalence of reticular pseudodrusen in age-related macular degeneration with newly diagnosed choroidal neovascularisation. Br. J. Ophthalmol. 91, 354–359 (2007).
Zweifel, S. A., Imamura, Y., Spaide, T. C., Fujiwara, T. & Spaide, R. F. Prevalence and significance of subretinal drusenoid deposits (reticular pseudodrusen) in age-related macular degeneration. Ophthalmology 117, 1775–1781 (2010).
Zhou, Q. et al. Pseudodrusen and Incidence of late age-related macular degeneration in fellow eyes in the comparison of age-related macular degeneration treatments trials. Ophthalmology 123, 1530–1540 (2016).
Lee, J. et al. Neovascularization in fellow eye of unilateral neovascular age-related macular degeneration according to different drusen types. Am. J. Ophthalmol. 208, 103–110 (2019).
Veerappan, M. et al. Optical coherence tomography reflective drusen substructures predict progression to geographic atrophy in age-related macular degeneration. Ophthalmology 123, 2554–2570 (2016).
VanderBeek, B. L. et al. Racial differences in age-related macular degeneration rates in the United States: a longitudinal analysis of a managed care network. Am. J. Ophthalmol. 152, 273–282 (2011).
Age-Related Eye Disease Study Research Group.A simplified severity scale for age-related macular degeneration. Arch. Ophthal. 123, 1570–1574 (2005).
Tonekaboni, S., Joshi, S., McCradden, M. D. & Goldenberg, A. M. What clinicians want: contextualizing explainable machine learning for clinical end use. Proc. Mach. Learn. Res. 106, 359–380 (2019).
Age-Related Eye Disease Study Research Group. The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs. Am. J. Ophthalmol. 132, 668–681 (2001).
Fragiotta, S., Rossi, T., Cutini, A., Grenga, P. L. & Vingolo, E. M. Predictive factors for development of neovascular age-related macular degeneration: a spectral-domain optical coherence tomography study. Retina 38, 245–252 (2018).
Schmidt-Erfurth, U. et al. Prediction of individual disease conversion in early AMD using artificial intelligence. Invest. Ophthalmol. Vis. Sci. 59, 3199–3208 (2018).
Abdelfattah, N. S. et al. Drusen volume as a predictor of disease progression in patients with late age-related macular degeneration in the fellow eye. Invest. Ophthalmol. Vis. Sci. 57, 1839–1846 (2016).
Folgar, F. A. et al. Drusen volume and retinal pigment epithelium abnormal thinning volume predict 2-year progression of age-related macular degeneration. Ophthalmology 123, 39–50 (2016).
NIHR Oxford Biomedical Research Centre. World’s First Gene Therapy Operation for Common Cause of Sight Loss Carried Out https://oxfordbrc.nihr.ac.uk/worlds-first-gene-therapy-operation-for-common-cause-of-sight-loss-carried-out/ (2019).
Dugel, P. U. et al. HAWK and HARRIER: phase 3, multicenter, randomized, double-masked trials of brolucizumab for neovascular age-related macular degeneration. Ophthalmology 127, 72–84 (2020).
Sahni, J. et al. Simultaneous Inhibition of angiopoietin-2 and vascular endothelial growth factor-a with faricimab in diabetic macular edema: BOULEVARD phase 2 randomized trial. Ophthalmology 126, 1155–1170 (2019).
Campochiaro, P. A. et al. The port delivery system with ranibizumab for neovascular age-related macular degeneration: results from the randomized phase 2 ladder clinical trial. Ophthalmology 126, 1141–1154 (2019).
Muether, P. S., Hermann, M. M., Koch, K. & Fauser, S. Delay between medical indication to anti-VEGF treatment in age-related macular degeneration can result in a loss of visual acuity. Graefes Arch. Clin. Exp. Ophthalmol. 249, 633–637 (2011).
Roisman, L. et al. Optical coherence tomography angiography of asymptomatic neovascularization in intermediate age-related macular degeneration. Ophthalmology 123, 1309–1319 (2016).
de Oliveira Dias, J. R. et al. Natural history of subclinical neovascularization in nonexudative age-related macular degeneration using swept-source oct angiography. Ophthalmology 125, 255–266 (2018).
Carnevali, A. et al. Natural history of treatment-naïve quiescent choroidal neovascularization in age-related macular degeneration using OCT angiography. Ophthalmol. Retina 2, 922–930 (2018).
Jager, R. D., Mieler, W. F. & Miller, J. W. Age-related macular degeneration. N. Engl. J. Med. 358, 2606–2617 (2008).
Babenko, B. et al. Predicting progression of age-related macular degeneration from fundus images using deep learning. Preprint at https://arxiv.org/abs/1904.05478 (2019).
Bogunovic, H. et al. Machine learning of the progression of intermediate age-related macular degeneration based on OCT Imaging. Invest. Ophthalmol. Vis. Sci. 58, BIO141–BIO150 (2017).
Russakoff, D. B., Lamin, A., Oakley, J. D., Dubis, A. M. & Sivaprasad, S. Deep learning for prediction of AMD progression: a pilot study. Invest. Ophthalmol. Vis. Sci. 60, 712–722 (2019).
Banerjee, I. et al. A deep-learning approach for prognosis of age-related macular degeneration disease using SD-OCT imaging biomarkers. Preprint at https://arxiv.org/abs/1902.10700 (2019).
Krause, J. et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology 125, 1264–1272 (2018).
Vander, J. F. Risk factors for the incidence of advanced age-related macular degeneration in the Age-Related Eye Disease Study (AREDS). Yearb. Ophthalmol. 2006, 119–121 (2006).
UK Information Commissioner’s Office. Anonymisation: Managing Data Protection Risk Code of Practice (2015).
De Fauw, J. et al. Automated analysis of retinal imaging using machine learning techniques for computer vision. F1000Res. 5, 1573 (2016).
Balaratnasingam, C. et al. Associations between retinal pigment epithelium and drusen volume changes during the lifecycle of large drusenoid pigment epithelial detachments. Invest. Ophthalmol. Vis. Sci. 57, 5479–5489 (2016).
Balaratnasingam, C. et al. Clinical characteristics, choroidal neovascularization, and predictors of visual outcomes in acquired vitelliform lesions. Am. J. Ophthalmol. 172, 28–38 (2016).
Lek, J. J. et al. Interpretation of subretinal fluid using OCT in intermediate age-related macular degeneration. Ophthalmol. Retina 2, 792–802 (2018).
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T. & Ronneberger, O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016 Vol. 9901, 424–432 (Springer, 2016).
Abadi, M. et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. Preprint at https://arxiv.org/abs/1603.04467 (2016).
Reynolds, M. et al. Open Sourcing Sonnet – A New Library for Constructing Neural Networks (DeepMind, accessed 26 July 2019); https://deepmind.com/blog/open-sourcing-sonnet/
Buchlovsky, P. et al. TF-Replicator: distributed machine learning for researchers. Preprint at https://arxiv.org/abs/1902.00465 (2019).
Curcio, C. A., Zanzottera, E. C., Ach, T., Balaratnasingam, C. & Freund, K. B. Activated retinal pigment epithelium, an optical coherence tomography biomarker for progression in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 58, BIO211–BIO226 (2017).
Christenbury, J. G. et al. and Age-Related Eye Disease Study 2 Ancillary Spectral Domain Optical Coherence Tomography Study Group. Progression of intermediate age-related macular degeneration with proliferation and inner retinal migration of hyperreflective foci. Ophthalmology 120, 1038–1045 (2013).
Folgar, F. A. et al. Spatial correlation between hyperpigmentary changes on color fundus photography and hyperreflective foci on SDOCT in intermediate AMD. Invest. Ophthalmol. Vis. Sci. 53, 4626–4633 (2012).
Age-Related Eye Disease Study Research Group. Risk factors associated with age-related macular degeneration. A case-control study in the age-related eye disease study: age-related eye disease study report number 3. Ophthalmology 107, 2224–2232 (2000).
Klein, R., Klein, B. E., Jensen, S. C. & Meuer, S. M. The five-year incidence and progression of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology 104, 7–21 (1997).
Bressler, S. B., Maguire, M. G., Bressler, N. M. & Fine, S. L. Relationship of drusen and abnormalities of the retinal pigment epithelium to the prognosis of neovascular macular degeneration. The Macular Photocoagulation Study Group. Arch. Ophthalmol. 108, 1442–1447 (1990).
Hinton, G., Vinyals, O. & Dean, J. Distilling the knowledge in a neural network. Preprint at https://arxiv.org/abs/1503.02531 (2015).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 4700–4708 (CVPR, 2017).
Xie, S., Sun, C., Huang, J., Tu, Z. & Murphy, K. In Proc. European Conference on Computer Vision 305–321 (Springer, 2018).
Kingma, D .P. & Ba, J. Adam: a method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2014).
Sadda, S. R. et al. Consensus definition for atrophy associated with age-related macular degeneration on OCT: classification of atrophy report 3. Ophthalmology 125, 537–548 (2018).
Acknowledgements
We thank the patients under the care of Moorfields Eye Hospital. We would also like to thank B. Romera-Paredes, O. Ronneberger, N. Tomasev, S. Blackwell, J. Schrouff, M. Chesus, C. Cooper, V. Cornelius, A. Khawaja, R. Ahamed, T. Corkett, R. Ogbe, Y. Ibitoye, M. Bawn, J. Besley, C. Meaden, C. Chorley, S. Rowley, A. Ahmad, K. Clancy, C. Semturs, A. Varadarajan, B. Babenko, I. Traynis, Y. Liu, L. Peng, N. Hammel, K. Blumer, K. Kavukcuoglu, S. Bouton, G. Corrado, E. Manna, A. C. Bird, W. Tucker, Y. Obadeyi, Z. Jessa, D. Sim, M. Natkunarajah and A. Jindal. P.A.K. is supported by an NIHR Clinician Scientist Award (no. NIHR-CS-2014-14-023). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. R.C. receives studentship support from the College of Optometrists, United Kingdom.
Author information
Authors and Affiliations
Contributions
R.C., P.T.K., D.K., D.H., M.S., T.B., J.R.L., P.A.K. and J.D.F. initiated the project. J.Y., R.C., T.S., H.A., M.L., J.H., K.F., G.M., J.R.L., P.A.K. and J.D.F. created the dataset. J.Y., T.S., J.W., H.A., M.W., J.D. and J.D.F. contributed to software engineering. J.Y., R.C., T.S., J.W., C.K., T.B., J.R.L., P.A.K. and J.D.F. analyzed the results. J.Y., R.C., T.S., J.W., A.O., C.K., H.A., T.B., J.R.L. and J.D.F. contributed to the overall experimental design. J.Y., J.W. and J.D.F. designed the model architectures. R.C., C.K., C.H., G.R., D.K., J.R.L. and P.A.K. contributed clinical expertise. J.Y., R.C., T.S., J.R.L., P.A.K. and J.D.F. contributed to subgroup analysis experiments. J.Y., R.C., C.K., T.S. and J.D.F. contributed to statistical analysis. J.Y., R.C., T.S., A.O., C.K., J.R.L., P.A.K. and J.D.F. contributed to the human evaluation. J.Y., R.C., T.S., J.W., C.K., M.L., J.H., M.W., J.D. and J.D.F. contributed to segmentation model improvements. J.Y., R.C., T.S., J.W., M.W., J.D. and J.D.F. contributed to experiments using segmentation data. J.Y., R.C., J.R.L., P.A.K. and J.D.F. contributed to literature reviews. J.Y., R.C., T.S., J.W., J.R.L., P.A.K. and J.D.F. contributed to false-positive analysis. A.O., H.A., C.M., T.B., J.R.L., P.A.K. and J.D.F. managed the project. J.Y., R.C., G.R., J.R.L., P.A.K. and J.D.F. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
P.A.K. and G.R. are paid contractors of DeepMind. P.A.K. has received speaker fees from Heidelberg Engineering, Topcon, Haag-Streit, Allergan, Novartis and Bayer. P.A.K. has served on advisory boards for Novartis and Bayer, and is an external consultant for DeepMind and Optos. M.L. received travel grants and a speaker fee from Bayer.
Additional information
Peer review information Michael Basson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Summary statistics and patient demographic data.
A breakdown of training (60%), validation (20%) and test (20%) datasets by unique patients and unique scans.
Extended Data Fig. 2 Dataset statistics.
a, Histogram of scans per unique eye (pre-conversion) in training/validation set and test set. b, Histogram of difference between conversion and injection date for fellow eyes in training and test set (n = 537, 214 have matching dates).
Extended Data Fig. 3 ROC curves for various time windows.
ROC curves for model predictions over time windows of a, 3 months, b, 6 months, c, 12 months, and d, 24 months. Note that the difference in AUC between 12 and 24 month predictions is not statistically significant (p-value = 0.54, two-sided permutation test).
Extended Data Fig. 4 Confusion matrices per expert and task.
Confusion matrices for the prediction decision for all 6 experts for the single scan and sequential scan tasks, and for the system at two chosen operating points. n = 1053 trials (380 unique patients).
Extended Data Fig. 5 Clinical expert metrics on the benchmark study.
a, Metrics for each expert for the single scan and sequential scan tasks. Intra-observer agreement was assessed using Fleiss’ Kappa. (PPV: positive predictive value, NPV: negative predictive value). b, Agreement between the clinical experts for the single and sequential tasks, measured using Fleiss’ Kappa. N = 1053 for both single and sequential task.
Extended Data Fig. 6 Aggregate volumes and volume change per 3 months before conversion for major ocular structures and abnormal tissues in patients who converted (n = 549 unique patients).
The box extends from the lower to upper quartile values of the data, with an orange line at the median. The whiskers show the 5th & 95th percentiles. For the left column, the statistics are calculated across patients, where patients with multiple scans per quarter are volumes averaged across these scans. For the right column the statistics for volume ch ange over 3 months were calculated on the difference for each patient between the mean volume for that quarter against the previous quarter. Volumes are calculated using the whole 2.3*6*6mm OCT volume.
Extended Data Fig. 7 Kaplan-Meier survival curves for full dataset and subgroups stratified by drusen stage and presence of HRF.
a, A Kaplan-Meier survival curve for fellow eye conversion to exAMD from baseline (defined as the first presentation of first eye conversion) in number of months, showing a little over 40% of patients converted during over 6 years of available follow up. The table shows number of eyes remaining at risk per month. b, The same plot for comparison with following plots. (c–g) Plots for varying amounts of drusen, showing increasing numbers of patient convert as drusen volume increases. Drusen size categories are calculated as quartiles. The same plots are shown for patients h, without and i, with geographic atrophy (GA), those j, without and k, with hyper-reflective foci (HRF), and those l, without and m, with fibrovascular pigment epithelial detachment (PED). In all plots the timeline is with reference to the first incidence of the feature in the eye.
Extended Data Fig. 8 Consort diagram.
Data labelling of the Moorfields Eye Hospital AMD dataset. Manual opt outs before data transfer are not included as none of the patients who manually opted out had digital OCT within the study dates.
Extended Data Fig. 9 Segmentation colour key and new hyperreflective foci class.
a, Colour key for 13 tissues and 3 artefacts segmented by the network. b, Left: Raw OCT input to the segmentation network. Right: Output of the retrained segmentation network. Three hyperreflective foci apparent in the intraretinal layers were successfully segmented in this B-scan (purple).
Extended Data Fig. 10 Deep learning system diagram.
Flow chart of the deep learning system including ensembling and TTA. Model inputs are shaped as trapezoids. Deep learning networks are shaped as rectangles. Model outputs are shaped as pointed rectangles. a, The segmentation network takes a raw OCT scan as input to generate a dense segmentation of the OCT which is then fed into a Diagnosis and referral network to obtain auxiliary task referral and diagnosis labels. b, The auxiliary labels along with either the raw OCT scan or dense segmentation are inputted into each exAMD prediction network across each cross validation fold group. Although the arrows apply to one fold group and instance, they generalise across all fold groups and instances. c, Ten TTA predictions are obtained from each instance. All TTA predictions are combined via averaging to obtain the final ensembled prediction. d, Architecture of a single block in our network. Green circles are convolution layers applied sequentially to the input of the previous layer. Each convolution has stride 1 and uses ReLU activation. Four convolutions are shown for demonstrative purposes but the number of convolutions and the kernels used for each will differ between blocks. Each convolution has a skip connection to the last orange node which concatenates all the intermediate and final activations along the channel dimension as the output.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and Tables 1–17
Rights and permissions
About this article
Cite this article
Yim, J., Chopra, R., Spitz, T. et al. Predicting conversion to wet age-related macular degeneration using deep learning. Nat Med 26, 892–899 (2020). https://doi.org/10.1038/s41591-020-0867-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-0867-7
This article is cited by
-
AI-based support for optical coherence tomography in age-related macular degeneration
International Journal of Retina and Vitreous (2024)
-
Behind the mask: a critical perspective on the ethical, moral, and legal implications of AI in ophthalmology
Graefe's Archive for Clinical and Experimental Ophthalmology (2024)
-
Optical coherence tomography imaging biomarkers associated with neovascular age-related macular degeneration: a systematic review
Eye (2023)
-
Predicting OCT biological marker localization from weak annotations
Scientific Reports (2023)
-
Deep learning for automated detection of neovascular leakage on ultra-widefield fluorescein angiography in diabetic retinopathy
Scientific Reports (2023)