Abstract
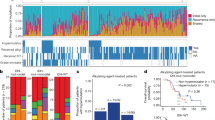
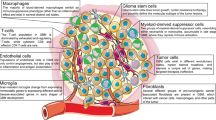
The failure to develop effective therapies for pediatric glioblastoma (pGBM) and diffuse intrinsic pontine glioma (DIPG) is in part due to their intrinsic heterogeneity. We aimed to quantitatively assess the extent to which this was present in these tumors through subclonal genomic analyses and to determine whether distinct tumor subpopulations may interact to promote tumorigenesis by generating subclonal patient-derived models in vitro and in vivo. Analysis of 142 sequenced tumors revealed multiple tumor subclones, spatially and temporally coexisting in a stable manner as observed by multiple sampling strategies. We isolated genotypically and phenotypically distinct subpopulations that we propose cooperate to enhance tumorigenicity and resistance to therapy. Inactivating mutations in the H4K20 histone methyltransferase KMT5B (SUV420H1), present in <1% of cells, abrogate DNA repair and confer increased invasion and migration on neighboring cells, in vitro and in vivo, through chemokine signaling and modulation of integrins. These data indicate that even rare tumor subpopulations may exert profound effects on tumorigenesis as a whole and may represent a new avenue for therapeutic development. Unraveling the mechanisms of subclonal diversity and communication in pGBM and DIPG will be an important step toward overcoming barriers to effective treatments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jones, C., Perryman, L. & Hargrave, D. Paediatric and adult malignant glioma: close relatives or distant cousins. Nat. Rev. Clin. Oncol. 9, 400–413 (2012).
Buczkowicz, P. et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat. Genet. 46, 451–456 (2014).
Fontebasso, A. M. et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat. Genet. 46, 462–466 (2014).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Taylor, K. R. et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat. Genet. 46, 457–461 (2014).
Wu, G. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 (2012).
Wu, G. et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat. Genet. 46, 444–450 (2014).
Sturm, D. et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat. Rev. Cancer 14, 92–107 (2014).
Gajjar, A. et al. Pediatric brain tumors: innovative genomic information is transforming the diagnostic and clinical landscape. J. Clin. Oncol. 33, 2986–2998 (2015).
Funato, K., Major, T., Lewis, P. W., Allis, C. D. & Tabar, V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346, 1529–1533 (2014).
Grasso, C. S. et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med. 21, 555–559 (2015).
Hashizume, R. et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat. Med. 20, 1394–1396 (2014).
Jones, C. & Baker, S. J. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat. Rev. Cancer 14, 651–661 (2014).
Prados, M. D. et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro-Oncol. 17, 1051–1063 (2015).
Sottoriva, A. et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA 110, 4009–4014 (2013).
Johnson, B. E. et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343, 189–193 (2014).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
McGranahan, N. & Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 27, 15–26 (2015).
Little, S. E. et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res. 72, 1614–1620 (2012).
Snuderl, M. et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 20, 810–817 (2011).
Szerlip, N. J. et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl. Acad. Sci. USA 109, 3041–3046 (2012).
Paugh, B. S. et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J. Clin. Oncol. 29, 3999–4006 (2011).
Puget, S. et al. Mesenchymal transition and PDGFRA amplification/mutation are key distinct oncogenic events in pediatric diffuse intrinsic pontine gliomas. PLoS One 7, e30313 (2012).
Nowak, M. A. Five rules for the evolution of cooperation. Science 314, 1560–1563 (2006).
Bolli, N. et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 5, 2997 (2014).
Van Loo, P. et al. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. USA 107, 16910–16915 (2010).
Andor, N., Harness, J. V., Müller, S., Mewes, H. W. & Petritsch, C. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics 30, 50–60 (2014).
Andor, N. et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2016).
Mackay, A. et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 32, 520–537.e525 (2017).
Caretti, V. et al. Subventricular spread of diffuse intrinsic pontine glioma. Acta Neuropathol. 128, 605–607 (2014).
Moniz, S. et al. Loss of WNK2 expression by promoter gene methylation occurs in adult gliomas and triggers Rac1-mediated tumour cell invasiveness. Hum. Mol. Genet. 22, 84–95 (2013).
Pollard, S. M. et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4, 568–580 (2009).
Monje, M. et al. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc. Natl. Acad. Sci. USA 108, 4453–4458 (2011).
Jørgensen, S., Schotta, G. & Sørensen, C. S. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 41, 2797–2806 (2013).
Jamal-Hanjani, M., Quezada, S. A., Larkin, J. & Swanton, C. Translational implications of tumor heterogeneity. Clin. Cancer Res. 21, 1258–1266 (2015).
Meyer, M. et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci. USA 112, 851–856 (2015).
Kreso, A. & Dick, J. E. Evolution of the cancer stem cell model. Cell Stem Cell 14, 275–291 (2014).
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011).
de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Morrissy, A. S. et al. Spatial heterogeneity in medulloblastoma. Nat. Genet. 49, 780–788 (2017).
Hoffman, L. M. et al. Spatial genomic heterogeneity in diffuse intrinsic pontine and midline high-grade glioma: implications for diagnostic biopsy and targeted therapeutics. Acta Neuropathol. Commun. 4, 1 (2016).
Nikbakht, H. et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat. Commun. 7, 11185 (2016).
Salloum, R. et al. Characterizing temporal genomic heterogeneity in pediatric high-grade gliomas. Acta Neuropathol. Commun. 5, 78 (2017).
Chapman, A. et al. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell. Rep. 8, 688–695 (2014).
Marusyk, A. et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514, 54–58 (2014).
Pai, C. C. et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 5, 4091 (2014).
Pfister, S. X. et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell. Rep. 7, 2006–2018 (2014).
Bouffet, E. et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 34, 2206–2211 (2016).
Castel, D. et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 130, 815–827 (2015).
International Cancer Genome Consortium PedBrain Tumor Project. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat. Med. 22, 1314–1320 (2016).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Vinci, M. et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 10, 29 (2012).
Vinci, M., Box, C. & Eccles, S. A. Three-dimensional (3D) tumor spheroid invasion assay. J. Vis. Exp. e52686, https://doi.org/10.3791/52686 (2015).
Vinci, M., Box, C., Zimmermann, M. & Eccles, S. A. Tumor spheroid-based migration assays for evaluation of therapeutic agents. Methods Mol. Biol. 986, 253–266 (2013).
Brough, R. et al. Functional viability profiles of breast cancer. Cancer Discov. 1, 260–273 (2011).
Postel-Vinay, S. et al. A high-throughput screen identifies PARP1/2 inhibitors as a potential therapy for ERCC1-deficient non-small cell lung cancer. Oncogene 32, 5377–5387 (2013).
Acknowledgements
We would like to thank L. Howell for confocal microcopy expertise, M. Lambros for technical advice, and H. Vogel for neuropathology expertise and assistance with autopsies at Stanford University. We are indebted to the multidisciplinary teams at the Royal Marsden Hospital, St George’s Hospital and Kings College Hospital for their continued assistance with prospective sample collection. Finally we thank the many children and families who contributed to this study through the donation of tumor tissue. This study makes use of data generated by the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project, C. Hawkins and the Hospital for Sick Children, and the McGill University-DKFZ Pediatric Brain Tumour Consortium. This work was supported by Cancer Research UK (grants C13468/A13982 and C13468/A23536), Abbie’s Army and the DIPG Collaborative, and the INSTINCT network funded by The Brain Tumour Charity, Great Ormond Street Children’s Charity and Children with Cancer UK. We further acknowledge funding from the Brainchild Foundation (Australia), Children’s Hospital Foundation Queensland, the Xarxa de Bancs de Tumors de Catalunya (XBTC) sponsored by Pla Director d’Oncologia de Catalunya, ISCIII-FEDER (CP13/00189), McKenna Claire Foundation, and the US National Institutes of Health (grants K08NS070926 and R01NS092597). We acknowledge the support of the Queensland Children’s Tumour Bank (QCTB) for provision of samples and clinical data. The QCTB is supported by the Children’s Hospital Foundation (Queensland) and the Brainchild Foundation. The authors acknowledge NHS funding to the NIHR Biomedical Research Centre at The Royal Marsden and the ICR and Experimental Cancer Medicines Centre (ECMC) funding.
Author information
Authors and Affiliations
Contributions
M.V., A.B., A.M. and C.J. conceived the study and wrote the manuscript with contributions from all authors. M.V., A.B., V.M., K.K., S.P., M.C., K.R.T., H.N.P., C.J.L., A.G., T.F. and D.C. carried out experiments. M.V., A.B., A.M. and C.J. analyzed data. L.V.M., E.Y.Q., W.J.I., A.S.M., H.-K.N., S.T., D.H.-B.B., N.E.-W., S.Z., S.V., H.C.M., L.R.B., A.J.M., S.A.-S., C.C., J.M., C.d.T., O.C., M.S., A.M.C., M.M. and A.M. prepared samples and provided clinical annotations. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Vinci, M., Burford, A., Molinari, V. et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat Med 24, 1204–1215 (2018). https://doi.org/10.1038/s41591-018-0086-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0086-7
This article is cited by
-
Joint analysis of mutational and transcriptional landscapes in human cancer reveals key perturbations during cancer evolution
Genome Biology (2024)
-
Evaluating cell culture reliability in pediatric brain tumor primary cells through DNA methylation profiling
npj Precision Oncology (2024)
-
Oncohistones and disrupted development in pediatric-type diffuse high-grade glioma
Cancer and Metastasis Reviews (2023)
-
Considerations for modelling diffuse high-grade gliomas and developing clinically relevant therapies
Cancer and Metastasis Reviews (2023)
-
Inhibition of exosome biogenesis affects cell motility in heterogeneous sub-populations of paediatric-type diffuse high-grade gliomas
Cell & Bioscience (2023)