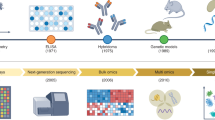
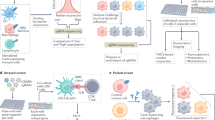
Abstract
The immune system comprises diverse specialized cell types that cooperate to defend the host against a wide range of pathogenic threats. Recent advancements in single-cell and spatial multi-omics technologies provide rich information about the molecular state of immune cells. Here, we review how the integration of single-cell and spatial multi-omics data with prior knowledge—gathered from decades of detailed biochemical studies—allows us to obtain functional insights, focusing on gene regulatory processes and cell–cell interactions. We present diverse applications in immunology and critically assess underlying assumptions and limitations. Finally, we offer a perspective on the ongoing technological and algorithmic developments that promise to get us closer to a systemic mechanistic understanding of the immune system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Arunachalam, P. S. et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369, 1210–1220 (2020).
Shen-Orr, S. S. & Furman, D. Variability in the immune system: of vaccine responses and immune states. Curr. Opin. Immunol. 25, 542–547 (2013).
Yofe, I., Dahan, R. & Amit, I. Single-cell genomic approaches for developing the next generation of immunotherapies. Nat. Med. 26, 171–177 (2020).
Banchereau, R., Cepika, A.-M. & Pascual, V. Systems approaches to human autoimmune diseases. Curr. Opin. Immunol. 25, 598–605 (2013).
Davis, M. M., Tato, C. M. & Furman, D. Systems immunology: just getting started. Nat. Immunol. 18, 725–732 (2017).
Germain, R. N., Meier-Schellersheim, M., Nita-Lazar, A. & Fraser, I. D. C. Systems biology in immunology: a computational modeling perspective. Annu. Rev. Immunol. 29, 527–585 (2011).
Ma, S. et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116 (2020). This work pioneered paired sequencing of chromatin accessibility and transcriptome in single cells (SHARE-seq).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). This work pioneered spatially resolved transcriptomics.
Regev, A. et al. The Human Cell Atlas. eLife https://doi.org/10.7554/eLife.27041 (2017).
Domínguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 376, eabl5197 (2022).
HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Martin, F. J. et al. Ensembl 2023. Nucleic Acids Res. 51, D933–D941 (2023).
UniProt Consortium. Uniprot: the universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2023).
Türei, D. et al. Integrated intra‐ and intercellular signaling knowledge for multicellular omics analysis. Mol. Syst. Biol. 17, e9923 (2021). This work introduces a comprehensive meta-resource that collects data from over a hundred databases, including, for example, protein interaction networks, ligand–receptor annotations and protein complex information.
Szklarczyk, D. et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44, D380–D384 (2016).
Garcia-Alonso, L., Holland, C. H., Ibrahim, M. M., Turei, D. & Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 29, 1363–1375 (2019).
Schubert, M. et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 (2018).
Armingol, E., Officer, A., Harismendy, O. & Lewis, N. E. Deciphering cell-cell interactions and communication from gene expression. Nat. Rev. Genet. 22, 71–88 (2021).
Vandereyken, K., Sifrim, A., Thienpont, B. & Voet, T. Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 24, 494–515 (2023).
Baysoy, A., Bai, Z., Satija, R. & Fan, R. The technological landscape and applications of single-cell multi-omics. Nat. Rev. Mol. Cell Biol. 24, 695–713 (2023).
Bonaguro, L. et al. A guide to systems-level immunomics. Nat. Immunol. 23, 1412–1423 (2022).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023).
Amezquita, R. A. et al. Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145 (2020).
Gene Ontology Consortium et al. The Gene Ontology knowledgebase in 2023. Genetics 224, iyad031 (2023).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Gillespie, M. et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 50, D687–D692 (2022).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Väremo, L., Nielsen, J. & Nookaew, I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 41, 4378–4391 (2013).
Badia-I-Mompel, P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv. 2, vbac016 (2022).
Hosokawa, H. & Rothenberg, E. V. How transcription factors drive choice of the T cell fate. Nat. Rev. Immunol. 21, 162–176 (2021).
Kawasaki, T. & Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 5, 461 (2014).
Saini, A., Ghoneim, H. E., Lio, C. -W. J., Collins, P. L. & Oltz, E. M. Gene regulatory circuits in innate and adaptive immune cells. Annu. Rev. Immunol. 40, 387–411 (2022).
Liu, Z.-P., Wu, C., Miao, H. & Wu, H. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database 2015, bav095 (2015).
Keenan, A. B. et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47, W212–W224 (2019).
Müller-Dott, S. et al. Expanding the coverage of regulons from high-confidence prior knowledge for accurate estimation of transcription factor activities. Nucleic Acids Res. 51, 10934–10949 (2023).
Holland, C. H. et al. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 21, 36 (2020).
Zeitlinger, J. Seven myths of how transcription factors read the cis-regulatory code. Curr. Opin. Syst. Biol. 23, 22–31 (2020).
Fiers, M. W. E. J. et al. Mapping gene regulatory networks from single-cell omics data. Brief. Funct. Genomics 17, 246–254 (2018).
Chen, S. & Mar, J. C. Evaluating methods of inferring gene regulatory networks highlights their lack of performance for single cell gene expression data. BMC Bioinformatics 19, 232 (2018).
Pratapa, A., Jalihal, A. P., Law, J. N., Bharadwaj, A. & Murali, T. M. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods 17, 147–154 (2020).
Badia-I-Mompel, P. et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 24, 739–754 (2023).
Wayman, J. A. et al. An atlas of gene regulatory networks for memory CD4+ T cells in youth and old age. Preprint at BioRxiv https://doi.org/10.1101/2023.03.07.531590 (2023).
Chowdhary, K. & Benoist, C. A variegated model of transcription factor function in the immune system. Trends Immunol. 44, 530–541 (2023).
Bravo González-Blas, C. et al. SCENIC+: single-cell multiomic inference of enhancers and gene regulatory networks. Nat. Methods 20, 1355–1367 (2023).
Kramer, B. A., Sarabia Del Castillo, J. & Pelkmans, L. Multimodal perception links cellular state to decision-making in single cells. Science 377, 642–648 (2022).
Weidemüller, P., Kholmatov, M., Petsalaki, E. & Zaugg, J. B. Transcription factors: bridge between cell signaling and gene regulation. Proteomics 21, e2000034 (2021).
Deribe, Y. L., Pawson, T. & Dikic, I. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672 (2010).
Lun, X. -K. & Bodenmiller, B. Profiling cell signaling networks at single-cell resolution. Mol. Cell. Proteom. 19, 744–756 (2020).
Dugourd, A. & Saez-Rodriguez, J. Footprint-based functional analysis of multiomic data. Curr. Opin. Syst. Biol. 15, 82–90 (2019).
Rydenfelt, M., Klinger, B., Klünemann, M. & Blüthgen, N. SPEED2: inferring upstream pathway activity from differential gene expression. Nucleic Acids Res. 48, W307–W312 (2020).
Jiang, P. et al. Systematic investigation of cytokine signaling activity at the tissue and single-cell levels. Nat. Methods 18, 1181–1191 (2021). This work introduced a comprehensive resource describing how cytokines affect gene expression, which can be used to infer cytokine activity from transcriptomics data.
Garrido-Rodriguez, M., Zirngibl, K., Ivanova, O., Lobentanzer, S. & Saez-Rodriguez, J. Integrating knowledge and omics to decipher mechanisms via large-scale models of signaling networks. Mol. Syst. Biol. 18, e11036 (2022).
Dimitrov, D. et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-seq data. Nat. Commun. 13, 3224 (2022).
Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018). This work introduced a popular tool for cell–cell communication analysis, which they used to systematically study immuneoregulatory mechanisms in the maternal–fetal interface.
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020). This work introduced a broadly used tools to analyze cell–cell communication analysis by integrating intracellular signaling networks.
Zhang, Y. et al. A T cell resilience model associated with response to immunotherapy in multiple tumor types. Nat. Med. 28, 1421–1431 (2022).
Wilk, A. J., Shalek, A. K., Holmes, S. & Blish, C. A. Comparative analysis of cell–cell communication at single-cell resolution. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01782-z (2023).
Raredon, M. S. B. et al. Comprehensive visualization of cell-cell interactions in single-cell and spatial transcriptomics with NICHES. Bioinformatics 39, btac775 (2023).
Zinner, M., Lukonin, I. & Liberali, P. Design principles of tissue organisation: how single cells coordinate across scales. Curr. Opin. Cell Biol. 67, 37–45 (2020).
Luthria, G., Lauffenburger, D. & Miller, M. A. Cell–cell communication networks in tissue: toward quantitatively linking structure with function. Curr. Opin. Syst. Biol. https://doi.org/10.1016/j.coisb.2021.05.002 (2021).
Graham, D. B. & Xavier, R. J. Conditioning of the immune system by the microbiome. Trends Immunol. 44, 499–511 (2023).
Zhang, B., Vogelzang, A. & Fagarasan, S. Secreted immune metabolites that mediate immune cell communication and function. Trends Immunol. 43, 990–1005 (2022).
Zheng, R. et al. MEBOCOST: metabolic cell-cell communication modeling by single cell transcriptome. Preprint at BioRxiv https://doi.org/10.1101/2022.05.30.494067 (2022).
Dimitrov, D. et al. LIANA+: an all-in-one cell-cell communication framework. Preprint at BioRxiv https://doi.org/10.1101/2023.08.19.553863 (2023).
Zhao, W., Johnston, K. G., Ren, H., Xu, X. & Nie, Q. Inferring neuron-neuron communications from single-cell transcriptomics through NeuronChat. Nat. Commun. 14, 1128 (2023).
Armingol, E., Larsen, R. O., Cequeira, M., Baghdassarian, H. & Lewis, N. E. Unraveling the coordinated dynamics of protein- and metabolite-mediated cell-cell communication. Preprint at BioRxiv https://doi.org/10.1101/2022.11.02.514917 (2022).
Cui, A. et al. Dictionary of immune responses to cytokines at single-cell resolution. Nature https://doi.org/10.1038/s41586-023-06816-9 (2023).
Liu, Z., Sun, D. & Wang, C. Evaluation of cell-cell interaction methods by integrating single-cell RNA sequencing data with spatial information. Genome Biol. 23, 218 (2022).
Browaeys, R. et al. MultiNicheNet: a flexible framework for differential cell-cell communication analysis from multi-sample multi-condition single-cell transcriptomics data. Preprint at bioRxiv https://doi.org/10.1101/2023.06.13.544751 (2023).
Armingol, E. et al. Context-aware deconvolution of cell–cell communication with Tensor-cell2cell. Nat. Commun. 13, 3665 (2022).
Chua, R. L. et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 38, 970–979 (2020).
Nasab, R. Z. et al. Deep learning in spatially resolved transcriptomics: a comprehensive technical view. Preprint at https://arxiv.org/abs/2210.04453 (2022).
Li, B. et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat. Methods 19, 662–670 (2022).
Dries, R. et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22, 78 (2021).
Garcia-Alonso, L. et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat. Genet. 53, 1698–1711 (2021).
Shao, X. et al. Knowledge-graph-based cell-cell communication inference for spatially resolved transcriptomic data with SpaTalk. Nat. Commun. 13, 4429 (2022).
Li, Z., Wang, T., Liu, P. & Huang, Y. SpatialDM: rapid identification of spatially co-expressed ligand-receptor reveals cell-cell communication patterns. Preprint at BioRxiv https://doi.org/10.1101/2022.08.19.504616 (2022).
Cang, Z. et al. Screening cell-cell communication in spatial transcriptomics via collective optimal transport. Nat. Methods 20, 218–228 (2023).
Frede, A. et al. B cell expansion hinders the stroma-epithelium regenerative cross talk during mucosal healing. Immunity 55, 2336–2351 (2022).
Lin, J.-R. et al. Multiplexed 3D atlas of state transitions and immune interaction in colorectal cancer. Cell 186, 363–381 (2023).
Bock, C. et al. High-content CRISPR screening. Nat. Rev. Methods Primers 2, 9 (2022).
Kustatscher, G. et al. Understudied proteins: opportunities and challenges for functional proteomics. Nat. Methods 19, 774–779 (2022).
Stražar, M. et al. HLA-II immunopeptidome profiling and deep learning reveal features of antigenicity to inform antigen discovery. Immunity 56, 1681–1698 (2023).
Huffman, R. G. et al. Prioritized mass spectrometry increases the depth, sensitivity and data completeness of single-cell proteomics. Nat. Methods 20, 714–722 (2023).
Makhmut, A. et al. A framework for ultra-low input spatial tissue proteomics. Cell Syst. https://doi.org/10.1016/j.cels.2023.10.003 (2023).
Rappez, L. et al. SpaceM reveals metabolic states of single cells. Nat. Methods 18, 799–805 (2021).
Vicari, M. et al. Spatial multimodal analysis of transcriptomes and metabolomes in tissues. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01937-y (2023).
Deng, Y. et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 609, 375–383 (2022).
Zhang, D. et al. Spatial epigenome-transcriptome co-profiling of mammalian tissues. Nature 616, 113–122 (2023).
Engblom, C. et al. Spatial transcriptomics of B cell and T cell receptors reveals lymphocyte clonal dynamics. Science 382, eadf8486 (2023).
Liu, Y. et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 41, 1405–1409 (2023).
Lötstedt, B., Stražar, M., Xavier, R., Regev, A. & Vickovic, S. Spatial host-microbiome sequencing reveals niches in the mouse gut. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01988-1 (2023).
Saarenpää, S. et al. Spatial metatranscriptomics resolves host-bacteria-fungi interactomes. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01979-2 (2023).
Udani, S. et al. Secretion encoded single-cell sequencing (SEC-seq) uncovers gene expression signatures associated with high VEGF-A secretion in mesenchymal stromal cells. Preprint at BioRxiv https://doi.org/10.1101/2023.01.07.523110 (2023).
Zhang, S. et al. Monitoring of cell-cell communication and contact history in mammals. Science 378, eabo5503 (2022).
Clark, I. C. et al. Barcoded viral tracing of single-cell interactions in central nervous system inflammation. Science 372, eabf1230 (2021).
Hor, J. L. & Germain, R. N. Intravital and high-content multiplex imaging of the immune system. Trends Cell Biol. 32, 406–420 (2022).
Chen, W. et al. Live-seq enables temporal transcriptomic recording of single cells. Nature 608, 733–740 (2022).
Kunes, R. Z. et al. Supervised discovery of interpretable gene programs from single-cell data. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01940-3 (2023).
Qoku, A. & Buettner, F. Encoding domain knowledge in multi-view latent variable models: a bayesian approach with structured sparsity. Preprint at https://arxiv.org/abs/2204.06242 (2022).
Lotfollahi, M. et al. Biologically informed deep learning to query gene programs in single-cell atlases. Nat. Cell Biol. 25, 337–350 (2023).
OpenAI. GPT-4 Technical Report. Preprint at https://arxiv.org/abs/2303.08774 (2023).
Cui, H., Wang, C., Maan, H. & Wang, B. scGPT: towards building a foundation model for single-cell multi-omics using generative AI. Preprint at BioRxiv https://doi.org/10.1101/2023.04.30.538439 (2023).
Theodoris, C. V. et al. Transfer learning enables predictions in network biology. Nature 618, 616–624 (2023).
Vodovotz, Y. et al. Solving immunology? Trends Immunol. 38, 116–127 (2017).
Handel, A., La Gruta, N. L. & Thomas, P. G. Simulation modelling for immunologists. Nat. Rev. Immunol. 20, 186–195 (2020).
Bachireddy, P. et al. Mapping the evolution of T cell states during response and resistance to adoptive cellular therapy. Cell Rep. 37, 109992 (2021).
Kuppe, C. et al. Spatial multi-omic map of human myocardial infarction. Nature 608, 766–777 (2022).
Kamal, A. et al. GRaNIE and GRaNPA: inference and evaluation of enhancer-mediated gene regulatory networks. Mol. Syst. Biol. 19, e11627 (2023).
Kamimoto, K. et al. Dissecting cell identity via network inference and in silico gene perturbation. Nature 614, 742–751 (2023). This work pioneered GRN inference from paired single-cell sequencing of chromatin accessibility and transcriptome and its use to predict perturbation effects.
Kwok, A. J. et al. Neutrophils and emergency granulopoiesis drive immune suppression and an extreme response endotype during sepsis. Nat. Immunol. 24, 767–779 (2023).
Han, A., Glanville, J., Hansmann, L. & Davis, M. M. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 (2014). This work pioneered paired sequencing of the transcriptome and TCR alpha/beta chains in single cells in a high-throughput manner.
Pai, J. A. & Satpathy, A. T. High-throughput and single-cell T cell receptor sequencing technologies. Nat. Methods 18, 881–892 (2021).
Valkiers, S. et al. Recent advances in T-cell receptor repertoire analysis: bridging the gap with multimodal single-cell RNA sequencing. ImmunoInformatics https://doi.org/10.1016/j.immuno.2022.100009 (2022).
Joglekar, A. V. & Li, G. T cell antigen discovery. Nat. Methods 18, 873–880 (2021).
Bradley, P. & Thomas, P. G. Using T cell receptor repertoires to understand the principles of adaptive immune recognition. Annu. Rev. Immunol. 37, 547–570 (2019).
Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018).
Glanville, J. et al. Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98 (2017).
Dash, P. et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547, 89–93 (2017).
Bassez, A. et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 27, 820–832 (2021).
Suo, C. et al. Dandelion uses the single-cell adaptive immune receptor repertoire to explore lymphocyte developmental origins. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01734-7 (2023).
Zhang, Z., Xiong, D., Wang, X., Liu, H. & Wang, T. Mapping the functional landscape of T cell receptor repertoires by single-T cell transcriptomics. Nat. Methods 18, 92–99 (2021).
Schattgen, S. A. et al. Integrating T cell receptor sequences and transcriptional profiles by clonotype neighbor graph analysis (CoNGA). Nat. Biotechnol. 40, 54–63 (2022).
An, Y., Drost, F., Theis, F., Schubert, B. & Lotfollahi, M. Integrating T-cell receptor and transcriptome for large-scale single-cell immune profiling analysis. Preprint at BioRxiv https://doi.org/10.1101/2021.06.24.449733 (2021).
Zhang, Z. et al. Interpreting the B-cell receptor repertoire with single-cell gene expression using Benisse. Nat. Mach. Intell. https://doi.org/10.1038/s42256-022-00492-6 (2022).
Shugay, M. et al. VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res. 46, D419–D427 (2018).
Hudson, D., Fernandes, R. A., Basham, M., Ogg, G. & Koohy, H. Can we predict T cell specificity with digital biology and machine learning? Nat. Rev. Immunol. 23, 511–521 (2023).
Osumi-Sutherland, D. et al. Cell type ontologies of the Human Cell Atlas. Nat. Cell Biol. 23, 1129–1135 (2021).
Lotfollahi, M. et al. Mapping single-cell data to reference atlases by transfer learning. Nat. Biotechnol. 40, 121–130 (2022).
Heimberg, G. et al. Scalable querying of human cell atlases via a foundational model reveals commonalities across fibrosis-associated macrophages. Preprint at BioRxiv https://doi.org/10.1101/2023.07.18.549537 (2023).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Jerby-Arnon, L. & Regev, A. DIALOGUE maps multicellular programs in tissue from single-cell or spatial transcriptomics data. Nat. Biotechnol. 40, 1467–1477 (2022).
Mitchel, J. et al. Tensor decomposition reveals coordinated multicellular patterns of transcriptional variation that distinguish and stratify disease individuals. Preprint at BioRxiv https://doi.org/10.1101/2022.02.16.480703 (2022).
Ramirez Flores, R. O., Lanzer, J. D., Dimitrov, D., Velten, B. & Saez-Rodriguez, J. Multicellular factor analysis of single-cell data for a tissue-centric understanding of disease. eLife 12, e93161 (2023).
Palla, G. et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods 19, 171–178 (2022).
Schapiro, D. et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387 (2018).
Chen, Z., Soifer, I., Hilton, H., Keren, L. & Jojic, V. Modeling multiplexed images with spatial-LDA reveals novel tissue microenvironments. J. Comput. Biol. 27, 1204–1218 (2020).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 (2018).
Schürch, C. M. et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359 (2020).
Wang, X. Q. et al. Spatial predictors of immunotherapy response in triple-negative breast cancer. Nature 621, 868–876 (2023).
Fischer, D. S., Schaar, A. C. & Theis, F. J. Modeling intercellular communication in tissues using spatial graphs of cells. Nat. Biotechnol. 41, 332–336 (2023).
Tanevski, J., Flores, R. O. R., Gabor, A., Schapiro, D. & Saez-Rodriguez, J. Explainable multiview framework for dissecting spatial relationships from highly multiplexed data. Genome Biol. 23, 97 (2022).
Wu, Z. et al. Graph deep learning for the characterization of tumour microenvironments from spatial protein profiles in tissue specimens. Nat. Biomed. Eng. 6, 1435–1448 (2022).
Sinha, S., Eisenhaber, B., Jensen, L. J., Kalbuaji, B. & Eisenhaber, F. Darkness in the human gene and protein function space: widely modest or absent illumination by the life science literature and the trend for fewer protein function discoveries since 2000. Proteomics 18, e1800093 (2018).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Ozaki, K. & Leonard, W. J. Cytokine and cytokine receptor pleiotropy and redundancy. J. Biol. Chem. 277, 29355–29358 (2002).
Korn, T. & Hiltensperger, M. Role of IL-6 in the commitment of T cell subsets. Cytokine 146, 155654 (2021).
Acknowledgements
P.S.L.S. has received funding from the Deutsche Forschungsgemeinschaft under grant agreement SPP 2395. D.D. is supported by the European Union’s Horizon 2020 research and innovation program (860329 Marie-Curie ITN ‘STRATEGY-CKD’). We thank R. O. R. Flores, P. Badia-i- Mompel, J. Tanevski, L. Küchenhoff, M. Garrido-Rodriguez, C. Lu and K. Mikulik for the helpful discussions.
Author information
Authors and Affiliations
Contributions
P.S.L.S., D.D. and J.S.-R. developed the concept based on starting ideas from J.S.-R. P.S.L.S. designed the figures. P.S.L.S., with assistance from D.D., wrote the original draft, with the supervision of J.S.-R. E.J.V. contributed immunological expertise. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.S.-R. reports funding from GSK, Pfizer and Sanofi and fees/honoraria from Travere Therapeutics, Stadapharm, Astex, Pfizer, Owkin and Grunenthal. E.J.V. has received research grants from F. Hoffmann-La Roche. All other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Jishnu Das and Yvan Saeys for their contribution to the peer review of this work. Primary Handling Editor: L. A. Dempsey, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Schäfer, P.S.L., Dimitrov, D., Villablanca, E.J. et al. Integrating single-cell multi-omics and prior biological knowledge for a functional characterization of the immune system. Nat Immunol 25, 405–417 (2024). https://doi.org/10.1038/s41590-024-01768-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-024-01768-2