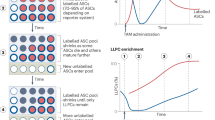
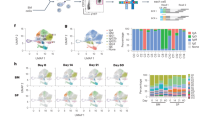
Abstract
Antibody-secreting plasma cells (PCs) are generated in secondary lymphoid organs but are reported to reside in an emerging range of anatomical sites. Analysis of the transcriptome of different tissue-resident (Tr)PC populations revealed that they each have their own transcriptional signature indicative of functional adaptation to the host tissue environment. In contrast to expectation, all TrPCs were extremely long-lived, regardless of their organ of residence, with longevity influenced by intrinsic factors like the immunoglobulin isotype. Analysis at single-cell resolution revealed that the bone marrow is unique in housing a compendium of PCs generated all over the body that retain aspects of the transcriptional program indicative of their tissue of origin. This study reveals that extreme longevity is an intrinsic property of TrPCs whose transcriptome is imprinted by signals received both at the site of induction and within the tissue of residence.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Tellier, J. & Nutt, S. L. Plasma cells: the programming of an antibody-secreting machine. Eur. J. Immunol. 49, 30–37 (2019).
Shi, W. et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 16, 663–673 (2015).
Wilson, R. P. et al. IgM plasma cells reside in healthy skin and accumulate with chronic inflammation. J. Invest. Dermatol. 139, 2477–2487 (2021).
Fitzpatrick, Z. et al. Gut-educated IgA plasma cells defend the meningeal venous sinuses. Nature 587, 472–476 (2020).
Palming, J. et al. Plasma cells and Fc receptors in human adipose tissue—lipogenic and anti-inflammatory effects of immunoglobulins on adipocytes. Biochem. Biophys. Res. Commun. 343, 43–48 (2006).
Sze, D. M., Toellner, K. M., García de Vinuesa, C., Taylor, D. R. & MacLennan, I. C. Intrinsic constraint on plasmablast growth and extrinsic limits of plasma cell survival. J. Exp. Med. 192, 813–821 (2000).
Cordero, H. et al. Intrathymic differentiation of natural antibody-producing plasma cells in human neonates. Nat. Commun. 12, 5761 (2021).
Manz, R. A., Thiel, A. & Radbruch, A. Lifetime of plasma cells in the bone marrow. Nature 388, 133–134 (1997).
Duan, M. et al. Understanding heterogeneity of human bone marrow plasma cell maturation and survival pathways by single-cell analyses. Cell Rep. 42, 112682 (2023).
Schoenhals, M. et al. Hypoxia favors the generation of human plasma cells. Cell Cycle 16, 1104–1117 (2017).
Kim, M., Qie, Y., Park, J. & Kim, C. H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20, 202–214 (2016).
Lam, W. Y. et al. Mitochondrial pyruvate import promotes long-term survival of antibody-secreting plasma cells. Immunity 45, 60–73 (2016).
Kallies, A. et al. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 200, 967–977 (2004).
Ramanan, D. et al. An immunologic mode of multigenerational transmission governs a gut treg setpoint. Cell 181, 1276–1290 (2020).
Fowler, T. et al. Divergence of transcriptional landscape occurs early in B cell activation. Epigenetics Chromatin 8, 20 (2015).
DiLillo, D. J. et al. Maintenance of long-lived plasma cells and serological memory despite mature and memory B cell depletion during CD20 immunotherapy in mice. J. Immunol. 180, 361–371 (2008).
Salmi, M. & Jalkanen, S. Lymphocyte homing to the gut: attraction, adhesion, and commitment. Immunol. Rev. 206, 100–113 (2005).
Van Spriel, A. B. et al. The tetraspanin CD37 orchestrates the α(4)β(1) integrin-Akt signaling axis and supports long-lived plasma cell survival. Sci. Signal 5, ra82 (2012).
Morteau, O. et al. An indispensable role for the chemokine receptor CCR10 in IgA antibody-secreting cell accumulation. J. Immunol. 181, 6309–6315 (2008).
Kim, S. V. et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 340, 1456–1459 (2013).
Xiong, L. et al. Ahr-Foxp3-RORγt axis controls gut homing of CD4+ T cells by regulating GPR15. Sci. Immunol. 5, eaaz7277 (2020).
Xu, A. Q., Barbosa, R. R. & Calado, D. P. Genetic timestamping of plasma cells in vivo reveals tissue-specific homeostatic population turnover. eLife 9, e59850 (2020).
Chevrier, S. et al. CD93 is required for maintenance of antibody secretion and persistence of plasma cells in the bone marrow niche. Proc. Natl Acad. Sci. USA 106, 3895–3900 (2009).
Chevrier, S. et al. The BTB-ZF transcription factor Zbtb20 is driven by Irf4 to promote plasma cell differentiation and longevity. J. Exp. Med. 211, 827–840 (2014).
Rozanski, C. H. et al. Sustained antibody responses depend on CD28 function in bone marrow-resident plasma cells. J. Exp. Med. 208, 1435–1446 (2011).
Wang, Y. & Bhattacharya, D. Adjuvant-specific regulation of long-term antibody responses by ZBTB20. J. Exp. Med. 211, 841–856 (2014).
Männe, C. et al. Salmonella SiiE prevents an efficient humoral immune memory by interfering with IgG+ plasma cell persistence in the bone marrow. Proc. Natl Acad. Sci. USA 116, 7425–7430 (2019).
Mina, M. J. et al. Measles virus infection diminishes preexisting antibodies that offer protection from other pathogens. Science 366, 599–606 (2019).
Penny, H. A. et al. Rhythmicity of intestinal IgA responses confers oscillatory commensal microbiota mutualism. Sci. Immunol. 7, eabk2541 (2022).
Gray, J. I. & Farber, D. L. Tissue-resident immune cells in humans. Annu. Rev. Immunol. 40, 195–220 (2022).
Vergani, S. et al. A self-sustaining layer of early-life-origin B cells drives steady-state IgA responses in the adult gut. Immunity 55, 1829–1842 (2022).
Yang, Y. et al. CTLA-4 expression by B-1a B cells is essential for immune tolerance. Nat. Commun. 12, 525 (2021).
Maseda, D. et al. Peritoneal cavity regulatory B cells (B10 cells) modulate IFN-γ+CD4+ T cell numbers during colitis development in mice. J. Immunol. 191, 2780–2795 (2013).
Wilmore, J. R. et al. IgA plasma cells are long-lived residents of gut and bone marrow that express isotype- and tissue-specific gene expression patterns. Front. Immunol. 12, 791095 (2021).
Higgins, B. W. et al. Isotype-specific plasma cells express divergent transcriptional programs. Proc. Natl Acad. Sci. USA 119, e2121260119 (2022).
Liu, X., Yao, J., Zhao, Y., Wang, J. & Qi, H. Heterogeneous plasma cells and long-lived subsets in response to immunization, autoantigen and microbiota. Nat. Immunol. 23, 1564–1576 (2022).
Price, M. J., Scharer, C. D., Kania, A. K., Randall, T. D. & Boss, J. M. Conserved epigenetic programming and enhanced heme metabolism drive memory B cell reactivation. J. Immunol. 206, 1493–1504 (2021).
Robinson, M. J. et al. Intrinsically determined turnover underlies broad heterogeneity in plasma-cell lifespan. Immunity 56, 1596–1612 (2023).
Koike, T. et al. Progressive differentiation toward the long-lived plasma cell compartment in the bone marrow. J. Exp. Med. 220, e20221717 (2022).
Watanabe, K. et al. Requirement for Runx proteins in IgA class switching acting downstream of TGF-β1 and retinoic acid signaling. J. Immunol. 184, 2785–2792 (2010).
Wellford, S. A. et al. Mucosal plasma cells are required to protect the upper airway and brain from infection. Immunity 55, 2118–2134 (2022).
Fenton, T. M. et al. Immune profiling of human gut-associated lymphoid tissue identifies a role for isolated lymphoid follicles in priming of region-specific immunity. Immunity 52, 557–570 (2020).
Masahata, K. et al. Generation of colonic IgA-secreting cells in the caecal patch. Nat. Commun. 5, 3704 (2014).
Kabashima, K. et al. Plasma cell S1P1 expression determines secondary lymphoid organ retention versus bone marrow tropism. J. Exp. Med. 203, 2683–2690 (2006).
Landsverk, O.J. B. et al. Antibody-secreting plasma cells persist for decades in human intestine. J. Exp. Med. 214, 309–317 (2017).
Robinson, M. J. et al. Long-lived plasma cells accumulate in the bone marrow at a constant rate from early in an immune response. Sci. Immunol. 7, eabm8389 (2022).
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Pinto, D. et al. A functional BCR in human IgA and IgM plasma cells. Blood 121, 4110–4114 (2013).
Tellier, J. et al. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat. Immunol. 17, 323–330 (2016).
Van de Perre, P. Transfer of antibody via mother’s milk. Vaccine 21, 3374–3376 (2003).
Shalapour, S. et al. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 551, 340–345 (2017).
Rojas, O. L. et al. Recirculating intestinal IgA-producing cells regulate neuroinflammation via IL-10. Cell 176, 610–624 (2019).
Chow, M. A., Lebwohl, B., Reilly, N. R. & Green, P. H. R. Immunoglobulin A deficiency in celiac disease. J. Clin. Gastroenterol. 46, 850–854 (2012).
Wong, R. et al. Affinity-restricted memory B cells dominate recall responses to heterologous flaviviruses. Immunity 53, 1078–1094 (2020).
Groom, J. R. et al. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity 37, 1091–1103 (2012).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
McCarthy, D. J. & Smyth, G. K. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 25, 765–771 (2009).
Nie, J., Tellier, J., Tarasova, I., Nutt, S. L. & Smyth, G. K. The T2T-CHM13 reference genome has more accurate sequences for immunoglobulin genes than GRCh38. Preprint at bioRxiv https://doi.org/10.1101/2023.05.24.542206 (2023).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Song, L. et al. TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods 18, 627–630 (2021).
Acknowledgements
We thank our animal technicians, S. Wilcox and the WEHI flow cytometry facility for excellent technical assistance. We thank C. Dawson (WEHI) for discussions and A. Cook, Brinda Vijaykumar and Christophe Benoist (Harvard Medical School) for their helpful assistance. This work was supported by the National Health and Medical Research Council Australia (1155342 and 1160830 to S.L.N. and 1058892 to G.K.S.), a Cancer Council Victoria grant in aid (to J.T. and S.L.N.), the Chan Zuckerberg Initiative (2021-237445 to G.K.S.) and in part by the NIH grant R24072073 to the ImmGen consortium.
Author information
Authors and Affiliations
Contributions
J.T. and S.L.N. conceptualized the study. J.T., I.T., J.N., C.S.S., P.L.F. and W.H.J.C. conducted the methodology. J.T., I.T., J.N. and P.L.F. conducted the investigation. J.T., I.T. and S.L.N. wrote the original draft of the manuscript. J.T., J.N., P.L.F., J.R.G., D.B., G.K.S. and S.L.N. conducted the review and editing of the manuscript. J.R.G., G.T.B., D.B. and S.L.N. arranged the resources. G.K.S. and S.L.N. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
Sana Biotechnology has licensed the intellectual property of D.B. and Washington University in St. Louis. Gilead Sciences has licensed the intellectual property of D.B. and Stanford University. D.B. is a cofounder of Clade Therapeutics. D.B. served on an advisory panel for GlaxoSmithKline. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: L. A. Dempsey, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Identifying TrPCs.
a, Gating strategy for the identification of ASCs in the different tissues. PB (GFPlo) and PC (GFPhi) were identified based on the expression of Blimp1-GFP and CD138 in the bone marrow (BM), Peyer’s patches (PP), mesenteric lymph nodes (mesLN), or mandibular LNs (manLN) and Blimp1-GFP and CD98 in the genital tract (GT), colon (Co), salivary gland (SG), thymus, mammary gland (MG) or visceral adipose tissue (VAT). b, Expression profile of the top 50 most downregulated genes in ASCs compared to B cells (left) or the top 50 upregulated genes in BMPCs compared to B cells. c, Gating strategy of the mouse BMPC subpopulations. PC (CD138hi Blimp1-GFPhi) were separated in two subpopulations based on the surface expression of IgA. d, Gating strategy of the PCs (CD38+ CD138+) from human BM. RNAseq data in b are from Fig. 1.
Extended Data Fig. 2 Differential expression of tissue specific genes.
Mean difference plots comparing gene expression in one tissue PC versus all the other tissue populations. Colored are the genes with a statistically significant (adj P value < 0.05) difference of expression between the two groups. RNAseq data in b are from Fig. 1.
Extended Data Fig. 3 Measuring the longevity of TrPCs.
a, Time stamping data to measure the persistence of Tomato+ ASC from the indicated tissues and populations to identify long-lived PCs. Data are from Fig. 3d and show the mean percentage of ASCs that are of Tomato+ over time. MG: mammary gland, PP: Peyer’s patches, BM: bone marrow, SI: small intestine, VAT: visceral adipose tissue, mesLN: mesenteric lymph nodes, manLN: mandibular LN, SG: salivary glands, GT: genital tract. Each dot is an individual mouse (n = 5). Unpaired t test with Welsh correction (alpha set at 0.05) between the 3-day and 10-month timepoints. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05. Each dot is an individual mouse. Data collected over >3 independent experiments. b, Quantification in TrPCs at 10 months post-tamoxifen treatment, separated by isotype. Each dot is an individual mouse. Each tissue is color-coded. Two-tailed unpaired t test to compare the isotype groups. **** P < 0.0001. c, Flow cytometry contour plots of ASCs from the BM of IgJCreERT2 Rosa26LSL-tdTomato mice 3 days after tamoxifen. Left: co-expression of Blimp1-GFP and the induced dtTomato reporter. Middle-right: gating strategy of IgA, IgG and IgM BMPCs.
Extended Data Fig. 4 Analysis of the BCR repertoire.
a, The stack column graphic shows the percentages of clones with each isotype for each single sample of ASCs from different tissues as indicated in the plot. Undetermined, indicates the clones which we were unable to link to an isotype. b-c, The boxplots show the value distribution of CDR3 lengths for clones with each isotype (b) and the value distribution of mutation rates for clones in ASCs from different tissues by isotype (c). A dot represents one single clone that has a outlier value. The central lines indicate the group median values, the top and bottom lines indicate the 75th and 25th percentiles, respectively, the whiskers represent minima and maxima within the 1.5× the interquartile range. d-e, Quantification of the Ighv repertoire diversity. d, The Gini-Simpson index was calculated for all TrPC populations. A value near 0 would indicate a monoclonal sample, while a value of 1 would be infinite diversity. e, Ighv gene usage percentage accumulation curves for clones in ASCs from different tissues by isotype. Each single line represents clones from a single tissue of the same isotype. The first dot of each line represents the percentage of the most abundant Ighv gene, the second dot represents the sum of the percentages of the first two most abundant Ighv genes, and so forth. A curve with a higher slope indicates a lower diversity.
Extended Data Fig. 5 Tissue specific gene expression in PCs.
a, Differential expression of the genes encoding protein modification enzymes in the TrPC populations. b, Differential expression of the molecular clock genes in the TrPC populations. In a and b the size of the bubble represents the average expression (in RPKM) and the color indicates the log2 fold change compared to BM. c, Surface expression of PDL1 by ASCs of the indicated tissues and spleen B cells. Representative flow cytometry histograms with MFI indicated in the left side. d, Surface expression of the chemokine receptor CXCR3 by colon lamina propria lymphocytes. Representative flow cytometry histograms with MFI indicated in the right side. e, Gating strategies of gut lamina propria immune populations. RNAseq data in a, b are from Fig. 1.
Extended Data Fig. 6 Combined murine spleen and bone marrow PC scRNAseq analysis.
a-g, Two dimensional tSNE plot of single cell transcriptome of murine (CD138+CD98hi) spleen and BMPCs. a, Cell clusters identified by Seurat’ s Louvain algorithm with resolution 0.3. b, Colors identify the tissue of origin (spleen or BM) of each cell. c, Expression of IgJ. d, Percentage of reads mapped to annotated immunoglobulin (Ig) genes out of all reads that were successfully mapped for each cell. Both heavy and light chain genes were included. e, Expression of Ighm (top), Igha (middle) and Ighg (bottom) isotypes. Ighg is the combination of Ighg1, Igh2b, Igh2c, Ighg3 gene expression. f, Expression of maturation-linked genes: Prg2, upregulated in PCs (top), Ssr2 downregulated in PCs (middle), Ccna2 expressed in cycling PBs (bottom). g, Restricted expression of Ccr10 (top) in IgA PCs, Camp (middle) in BMPCs, Chst1 (bottom) in IgM/IgG PCs.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tellier, J., Tarasova, I., Nie, J. et al. Unraveling the diversity and functions of tissue-resident plasma cells. Nat Immunol 25, 330–342 (2024). https://doi.org/10.1038/s41590-023-01712-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-023-01712-w
This article is cited by
-
Tissues of origin matter to plasma cell longevity
Nature Immunology (2024)