Abstract
In cancer and infections, self-renewing stem-like CD8+ T cells mediate the response of immunotherapies and replenish terminally exhausted T cells and effector-like T cells. However, the programs governing the lineage choice in chimeric antigen receptor (CAR) T cells are unclear. Here, by simultaneously profiling single-cell chromatin accessibility and transcriptome in the same CAR T cells, we identified heterogeneous chromatin states within CD8+ T cell subsets that foreshadowed transcriptional changes and were primed for regulation by distinct transcription factors. Transcription factors that controlled each CD8+ T cell subset were regulated by high numbers of enhancers and positioned as hubs of gene networks. FOXP1, a hub in the stem-like network, promoted expansion and stemness of CAR T cells and limited excessive effector differentiation. In the effector network, KLF2 enhanced effector CD8+ T cell differentiation and prevented terminal exhaustion. Thus, we identified gene networks and hub transcription factors that controlled the differentiation of stem-like CD8+ CAR T cells into effector or exhausted CD8+ CAR T cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated to support this study are available within the paper. The scRNA-seq, scATAC+RNA-seq and ChIP-seq data have been deposited at Gene Expression Omnibus (GSE202543). Source data are provided with this paper.
Code availability
Custom code used for scRNA-seq, scATAC+RNA-seq and ChIP-seq data analyses is available from the corresponding authors upon request.
References
Wu, T. et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, eaai8593 (2016).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016).
He, R. et al. Follicular CXCR5- expressing CD8+ T cells curtail chronic viral infection. Nature 537, 412–428 (2016).
Utzschneider, D. T. et al. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016).
Leong, Y. A. et al. CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17, 1187–1196 (2016).
Siddiqui, I. et al. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195–211 (2019).
Kurtulus, S. et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1−CD8+ tumor-infiltrating T cells. Immunity 50, 181–194 (2019).
Brummelman, J. et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J. Exp. Med. 215, 2520–2535 (2018).
Burger, M. L. et al. Antigen dominance hierarchies shape TCF1+ progenitor CD8 T cell phenotypes in tumors. Cell 184, 4996–5014 (2021).
Connolly, K. A. et al. A reservoir of stem-like CD8+ T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Sci. Immunol. 6, eabg7836 (2021).
Schenkel, J. M. et al. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1+ CD8+ T cells in tumor-draining lymph nodes. Immunity 54, 2338–2353 (2021).
Eberhardt, C. S. et al. Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature 597, 279–284 (2021).
Yao, C. et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20, 890–901 (2019).
Yao, C. et al. BACH2 enforces the transcriptional and epigenetic programs of stem-like CD8+ T cells. Nat. Immunol. 22, 370–380 (2021).
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013 (2018).
Miller, B. C. et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Chen, G. M. et al. Integrative bulk and single-cell profiling of premanufacture T-cell populations reveals factors mediating long-term persistence of CAR T-cell therapy. Cancer Discov. 11, 2186–2199 (2021).
Fraietta, J. A. et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 24, 563–571 (2018).
Deng, Q. et al. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat. Med. 26, 1878–1887 (2020).
Zander, R. et al. CD4+ T cell help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity 51, 1028–1042 (2019).
Hudson, W. H. et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+stem-like CD8+ T cells during chronic infection. Immunity 51, 1043–1058 (2019).
Beltra, J. C. et al. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52, 825–841 (2020).
Sandu, I. et al. Landscape of exhausted virus-specific CD8 T cells in chronic LCMV infection. Cell Rep. 32, 108078 (2020).
Yan, Y. et al. CX3CR1 identifies PD-1 therapy-responsive CD8+ T cells that withstand chemotherapy during cancer chemoimmunotherapy. JCI Insight 3, e97828 (2018).
Yang, Y. et al. TCR engagement negatively affects CD8 but not CD4 CAR T cell expansion and leukemic clearance. Sci. Transl. Med. 9, eaag1209 (2017).
Hu, B. et al. Augmentation of antitumor immunity by human and mouse CAR T cells secreting IL-18. Cell Rep. 20, 3025–3033 (2017).
Wilson, T. L. et al. Common trajectories of highly effective CD19-specific CAR T cells identified by endogenous T-cell teceptor lineages. Cancer Discov. 12, 2098–2119 (2022).
Good, C. R. et al. An NK-like CAR T cell transition in CAR T cell dysfunction. Cell 184, 6081–6100 (2021).
Giles, J. R. et al. Shared and distinct biological circuits in effector, memory and exhausted CD8+ T cells revealed by temporal single-cell transcriptomics and epigenetics. Nat. Immunol. 23, 1600–1613 (2022).
Singer, M. et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell 166, 1500–1511 (2016).
Andreatta, M. et al. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 12, 2965 (2021).
Salmon, A. J. et al. BHLHE40 regulates the T-cell effector function required for tumor microenvironment remodeling and immune checkpoint-therapy efficacy. Cancer Immunol. Res. 10, 597–611 (2022).
Dominguez, C. X. et al. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 212, 2041–2056 (2015).
Omilusik, K. D. et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212, 2027–2039 (2015).
Kasmani, M. Y. et al. Clonal lineage tracing reveals mechanisms skewing CD8+ T cell fate decisions in chronic infection. J. Exp. Med. 220, e20220679 (2023).
Ma, S. et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116 (2020).
Alfei, F. et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019).
Khan, O. et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019).
Scott, A. C. et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019).
Stephen, T. L. et al. SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells. Immunity 46, 51–64 (2017).
Stephen, T. L. et al. Transforming growth factor beta-mediated suppression of antitumor T cells requires FoxP1 transcription factor expression. Immunity 41, 427–439 (2014).
Paley, M. A. et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012).
Backer, R. A. et al. A central role for Notch in effector CD8+ T cell differentiation. Nat. Immunol. 15, 1143–1151 (2014).
Wang, D. et al. The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48, 659–674 (2018).
Shan, Q. et al. The transcription factor Runx3 guards cytotoxic CD8+ effector T cells against deviation towards follicular helper T cell lineage. Nat. Immunol. 18, 931–939 (2017).
Milner, J. J. et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017).
Omilusik, K. D. et al. Sustained Id2 regulation of E proteins is required for terminal differentiation of effector CD8+ T cells. J. Exp. Med. 215, 773–783 (2018).
Kurachi, M. et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15, 373–383 (2014).
Chen, Y. et al. BATF regulates progenitor to cytolytic effector CD8+ T cell transition during chronic viral infection. Nat. Immunol. 22, 996–1007 (2021).
Seo, H. et al. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol. 22, 983–995 (2021).
Riegel, D. et al. Integrated single-cell profiling dissects cell-state-specific enhancer landscapes of human tumor-infiltrating CD8+ T cells. Mol. Cell 83, 622–636 (2023).
Feng, X. et al. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat. Immunol. 12, 544–550 (2011).
Carlson, C. M. et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442, 299–302 (2006).
Jacoby, E. et al. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat. Commun. 7, 12320 (2016).
Kochenderfer, J. N., Yu, Z., Frasheri, D., Restifo, N. P. & Rosenberg, S. A. Adoptive transfer of syngeneic T cells transduced with a chimeric antigen receptor that recognizes murine CD19 can eradicate lymphoma and normal B cells. Blood 116, 3875–3886 (2010).
Schmidl, C., Rendeiro, A. F., Sheffield, N. C. & Bock, C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015).
Acknowledgements
We thank A. Mobley (UTSW), A. Guzman (UTSW), P. Raj (UTSW), G. Hedlund (CU), T. Kupfer (CU), D. Diener (CU) and O. Cho (CU) for their outstanding technical support. MSCV-mCD19-CD28-Z-tEGFR plasmid and E2A-PBX1 cell line were generated by T. Fry and K. Degolier (CU). B16-CD19 cell line was a gift from C. June (UPenn). This research was supported in part by grants from the National Institutes of Health (AI158294, AG083398, AG056524), a CRI CLIP, a V Scholar Award and an AFAR Grant for Junior Faculty to T. Wu; and a grant from the National Institutes of Health (AI154450), a grant from CPRIT (RR210035) and a grant from Department of Defense (HT94252310801) to C. Yao. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
T. Wu and C.Y. designed the experiments. Z.Z., G.L., X.-L.T., H.W., Y.L., W.S., K.Y., S.H., K.D., C.L., C.Y. and T. Wu performed the experiments. Z.Z., G.L., H.H., Q.Z., T.F., T. Wang, C.Y. and T. Wu analyzed and interpreted the results. T. Wu and C.Y. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
T. Wang is a co-founder of NightStar Biotechnologies. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Chiara Bonini and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ioana Visan, in collaboration with the Nature Immunology editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 CAR T cells in B cell ALL and melanoma show different single-cell transcriptomes.
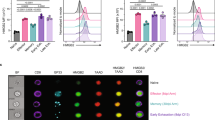
a, Upper left: Leukemia burden in E2A-PBX1-bearing mice that were untreated or treated with 106 CD19 CAR T cells (n = 5 mice per group). Upper right: Tumor burden in B16-CD19-bearing mice that were untreated (n = 5 mice) or treated (n = 10 mice) with 5 × 106 CD19 CAR T cells. Lower panel: Gating strategy for sorting CAR T cells (huEGFR+CD45.1+) from treated mice as in Fig. 1a. b, scRNA-seq profiles from different individual mice (replicates) in the same group are shown in separate UMAP plots. scRNA-seq is described in Fig. 1a. c, scRNA-seq profiles from different individual mice (replicates) in the same group are combined in the same UMAP plot. Cells are color-coded by the cluster defined in Fig. 1a. d, Single-cell expression of selected genes in CD8+ CAR T cells from mice with E2A-PBX1 as described in Fig. 1e is shown in UMAP plots. The mRNA level is color-coded. e, Single-cell expression of selected genes in CD8+ CAR T cells infiltrating B16-CD19 as described in Fig. 1h is shown in UMAP plots. The mRNA level is color-coded. f, Projection of cells in clusters 2, 4, 5 and 7 (black) in Fig. 1a to the reference map of ProjecTILs. Color codes of T cell subsets are based on the original ProjecTILs paper. g, A volcano plot comparing the transcriptome between CD8+ CAR T cells responding to E2A-PBX1 and those responding to B16-CD19. Statistical significance in a is determined by a two-sided Student’s t-test. Data in a are representative of two independent experiments. Circles in a left panel represent individual mice. Data in a right panel represent Mean ± SEM.
Extended Data Fig. 2 scATAC+RNA-seq identifies heterogeneous chromatin states in CD8+ CAR T cells in B cell ALL.
a, Percentages of mRNA reads from the scATAC+RNA-seq experiment (Fig. 2) or scRNA-seq experiment (Fig. 1) in the exonic, intronic or intergenic regions. b, A UMAP plot of CD8+ CAR T cells based on the RNA portion of scATAC+RNA-seq as in Fig. 2. Cells are color-coded based on cluster IDs defined by the transcriptome. c, A heatmap of differentially expressed genes in the Ts-like, Teff-like, ISG, and proliferating subsets defined in b. d, Gene-set enrichment analysis confirms the enrichment of Ts-like, Teff-like, proliferating or ISG signature defined in Fig. 1 in the corresponding clusters defined in Fig. 2a. The normalized enrichment score (NES) and adjusted P-value (adj P) are shown. Benjamini–Hochberg is used to calculate adjusted P values. e, Single-cell expression of ZEB2 shown in a UMAP generated from the ATAC portion of scATAC+RNA-seq described in Fig. 2.
Extended Data Fig. 3 scATAC+RNA-seq reveals transcriptional programs controlling melanoma-infiltrating CAR T cells.
a, A UMAP plot of tumor-infiltrating CD8+ CAR T cells based on the RNA-portion of the scATAC+RNA-seq data as in Fig. 3. Cells are color-coded by the cluster defined by the transcriptome. b, A heatmap of differentially expressed genes in each cluster defined in a. c, Gene-set enrichment analysis confirms the enrichment of Tex-like, Nr4a1+ Tex-like, ISG, proliferating or Klf2+ Tex-like signature defined in Fig. 1 in the corresponding clusters defined in Fig. 3a. The normalized enrichment score (NES) and adjusted P-value (adj P) are shown. Benjamini–Hochberg is used to calculate adjusted P values. d, Upper panel: A volcano plot showing DA peaks between B16_ATAC_2 and B16_ATAC_1 defined in Fig. 3a. Lower panel: A volcano plot showing DA peaks between B16_ATAC_4 and B16_ATAC_0 defined in Fig. 3a. e, Violin plots show the single-cell enrichment of short-lived effector cell signature (SLEC) in ATAC-based clusters defined in Fig. 3a. f, Merged scATAC-seq profiles of CD8+ CAR T cells in E2A-PBX1 (Fig. 2) and B16-CD19 (Fig. 3) shown by sample types (left) or cluster IDs (right). g, Chromatin accessibility profiles at selected genes in CD8+ CAR T cells in E2A-PBX1 (red) and B16-CD19 (blue) as in f. h, Motif enrichment of selected transcription factors in merged scATAC-seq profiles of CD8+ CAR T cells in E2A-PBX1 and B16-CD19 as in f.
Extended Data Fig. 4 scATAC+RNA-seq identifies DORCs and gene regulatory networks in CAR T cells.
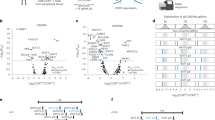
a, Peak-gene links are identified with scATAC+RNA-seq data in Figs. 2 and 3. The number of peak-gene links plotted against the distance to linked gene TSS in CD8+ CAR T cells from mice inoculated with E2A-PBX1 (left) or B16-CD19 (right). b, The distribution of the number of peaks linked to each gene (left) or the number of genes linked to each peak (right) as in a. c, Genomic loci of two representative DORCs, Nr4a1 and Tox, in CD8+ CAR T cells against B16-CD19. Chromatin accessibility and mRNA level (violin plots) of indicated genes in each CAR T cell subset defined by scATAC-seq as in Fig. 3a are shown. d,e, Heatmaps illustrating the expression of DORC-associated genes in each subset identified in CD8+ CAR T cells against E2A-PBX1 (d) as in Extended Data Fig. 2b or B16-CD19 (e) as in Extended Data Fig. 3a. f, g, Scatter plots showing all association between transcription factors and DORC-associated genes in CD8+ CAR T cells against E2A-PBX1 (f) or B16-CD19 (g) defined in Fig. 4a. For each transcription factor-DORC pair, transcription factor motif enrichment in the DORC is plotted against the correlation between the mRNA level of the enriched transcription factor and the chromatin accessibility of the DORC. h, Transcription factors with regulation score >1 in CD8+ CAR T cells from mice inoculated with E2A-PBX1 or B16-CD19. i,j, Gene regulatory networks in CD8+ CAR T cells from mice inoculated with E2A-PBX1 (i) or B16-CD19 (j) are constructed using transcription factor to DORC-containing gene associations. k, Gene regulatory network in the Tex-like subset from B16-CD19. l, Enrichment of hub transcription factors from the Ts-like subset, Teff-like subset or Tex-like subset in the TCF7 regulon in B cell ALL patients treated with CD19 CAR T cells23. One-sided Fisher’s exact test was used to calculate the P Value.
Extended Data Fig. 5 FOXP1 KO CD8+ CAR T cells show a defective antitumor immune response and altered molecular program.
a, The experiment was set up as Fig. 5. Frequencies of CD62L−CX3CR1+ WT and FOXP1 KO CD8+ CAR T cells are shown in bar graphs (n = 5 mice per group). b,c, Tumor growth (b) and survival (c) of C57BL6 mice that were inoculated with B16-CD19 tumor and treated with 106 WT (n = 10 mice) or FOXP1 KO (n = 11 mice) CD8+ CAR T cells at day 10 post-inoculation. d,e, Representative flow cytometry plots (d) and numbers (e) of WT (n = 6 mice) and FOXP1 KO (n = 7 mice) CD8+ CAR T cells in B16-CD19 tumors at day 8 after CAR T cell infusion as in b. f, The scATAC+RNA-seq data was generated as described in Fig. 5. Chromatin accessibility and expression of Bach2 and Klf2 in WT and FOXP1 KO CD8+ CAR T cells are shown. g, Motif enrichment of FOXP1 ChIP-seq peaks in CD8+ T cells. h, Distribution of FOXP1 ChIP-seq peaks in genomic features. i, FOXP1 binding sites at selected genes. j, A Venn diagram of FOXP1−bound genes in g, differentially expressed (DE) genes in Fig. 5j and genes with differentially accessible (DA) peaks in Fig. 5l. Statistical significance in a,b,e is determined by a two-sided Student’s t-test. Statistical significance in c is determined by log rank test. Data in a–e are representative of two independent experiments. Circles in a,e represent individual mice. Data in a,b,e represent the Mean ± SEM.
Extended Data Fig. 6 KLF2 is required for the differentiation of Teff-like CD8+ CAR T cells.
a,b, Representative flow cytometry plots (a) and numbers (b) of CD8+ CAR T cells in B16-CD19 tumors from C57BL6 mice that were inoculated with B16-CD19, treated with 106 WT (n = 7 mice) or KLF2 KO (n = 6 mice) CD8+ CAR T cells at day 10 post-inoculation and analyzed at day 7 after CAR T cell infusion. c,d, Expression of granzyme B (c) and PD1 (d) in B16-CD19-infiltrating WT (n = 7 mice) and KLF2 KO (n = 6 mice) CD8+ CAR T cells as in a. e, Killing of B16-CD19 cells that were co-cultured with WT or KLF2 KO (n = 3 independent cultures per group) CD8+ CAR T cells at different effector-to-target ratios. f,g, Tumor growth (f) and survival (g) of B16-CD19-bearing mice treated with WT or KLF2 KO (n = 7 mice per group) CD8+ CAR T cells as in a. h, The scATAC+RNA-seq data was generated as described in Fig. 6. Chromatin accessibility and expression of selected genes in WT and KLF2 KO CD8+ CAR T cells are shown. Boxes highlight DA peaks. i, Transcription factor motif enrichment at KLF2 ChIP-seq peaks. j, Distribution of KLF2 ChIP-seq peaks in genomic features. k, KLF2 binding at selected genes. l, A Venn diagram of KLF2-bound genes in i, differentially expressed (DE) genes in Fig. 6h and genes with differentially accessible (DA) peaks in Fig. 6n. Statistical significance in b-f is determined by a two-sided Student’s t-test. Statistical significance in g is determined by log rank test. Data in a–g are representative of two independent experiments. Circles in b-d represent individual mice. Data in b-f represent the Mean ± SEM.
Supplementary information
Source data
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, Z., Lou, G., Teng, XL. et al. FOXP1 and KLF2 reciprocally regulate checkpoints of stem-like to effector transition in CAR T cells. Nat Immunol 25, 117–128 (2024). https://doi.org/10.1038/s41590-023-01685-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-023-01685-w
This article is cited by
-
Epigenetic checkpoints regulate the fate and function of CAR-T cells
Nature Immunology (2024)