Abstract
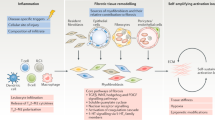
Fibrosis, defined by the excess deposition of structural and matricellular proteins in the extracellular space, underlies tissue dysfunction in multiple chronic diseases. Approved antifibrotics have proven modest in efficacy, and the immune compartment remains, for the most part, an untapped therapeutic opportunity. Recent single-cell analyses have interrogated human fibrotic tissues, including immune cells. These studies have revealed a conserved profile of scar-associated macrophages, which localize to the fibrotic niche and interact with mesenchymal cells that produce pathological extracellular matrix. Here we review recent advances in the understanding of the fibrotic microenvironment in human diseases, with a focus on immune cell profiles and functional immune–stromal interactions. We also discuss the key role of the immune system in mediating fibrosis regression and highlight avenues for future study to elucidate potential approaches to targeting inflammatory cells in fibrotic disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wynn, T. A. Fibrotic disease and the TH1/TH2 paradigm. Nat. Rev. Immunol. 4, 583–594 (2004).
Taylor, R. S. et al. Association between fibrosis stage and outcomes of patients with nonalcoholic fatty liver disease: a systematic review and meta-analysis. Gastroenterology 158, 1611–1625 (2020).
Moon, A. M., Singal, A. G. & Tapper, E. B. Contemporary epidemiology of chronic liver disease and cirrhosis. Clin. Gastroenterol. Hepatol. 18, 2650–2666 (2020).
Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019). scRNA-seq study of healthy versus fibrotic human liver, defining the transcriptional profile of SAMacs and studying ligand–receptor interactions in the fibrotic niche.
Buonomo, E. L. et al. Liver stromal cells restrict macrophage maturation and stromal IL-6 limits the differentiation of cirrhosis-linked macrophages. J. Hepatol. 76, 1127–1137 (2022).
Hendrikx, T. et al. Soluble TREM2 levels reflect the recruitment and expansion of TREM2+ macrophages that localize to fibrotic areas and limit NASH. J. Hepatol. 77, 1373–1385 (2022).
Remmerie, A. et al. Osteopontin expression identifies a subset of recruited macrophages distinct from kupffer cells in the fatty liver. Immunity 53, 641–657 (2020).
Xiong, X. et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 75, 644–660 (2019).
Daemen, S. et al. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep. 34, 108626 (2021).
Fabre, T. et al. Identification of a broadly fibrogenic macrophage subset induced by type 3 inflammation. Sci. Immunol. 8, eadd8945 (2023). Integrated analysis of lung and liver scRNA-seq data from human and mouse, defining the conserved features of SAMacs across organs. Highlights the role of GM-CSF, IL-17A and TGFβ in SAMac differentiation.
Guilliams, M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396 (2022).
Govaere, O. et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci. Transl. Med. 12, eaba4448 (2020).
Poch, T. et al. Single-cell atlas of hepatic T cells reveals expansion of liver-resident naive-like CD4+ T cells in primary sclerosing cholangitis. J. Hepatol. 75, 414–423 (2021).
Rau, M. et al. Progression from nonalcoholic fatty liver to nonalcoholic steatohepatitis is marked by a higher frequency of TH17 cells in the liver and an increased TH17/resting regulatory T cell ratio in peripheral blood and in the liver. J. Immunol. 196, 97–105 (2016).
Meng, F. et al. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology 143, 765–776 (2012).
Dudek, M. et al. Auto-aggressive CXCR6+ CD8 T cells cause liver immune pathology in NASH. Nature 592, 444–449 (2021).
Pallett, L. J. et al. Tissue CD14+CD8+ T cells reprogrammed by myeloid cells and modulated by LPS. Nature 614, 334–342 (2023).
Wang, S. et al. An autocrine signaling circuit in hepatic stellate cells underlies advanced fibrosis in nonalcoholic steatohepatitis. Sci. Transl. Med. 15, eadd3949 (2023).
Wallace, S. J., Tacke, F., Schwabe, R. F. & Henderson, N. C. Understanding the cellular interactome of non-alcoholic fatty liver disease. JHEP Rep. 4, 100524 (2022).
Ramachandran, P., Matchett, K. P., Dobie, R., Wilson-Kanamori, J. R. & Henderson, N. C. Single-cell technologies in hepatology: new insights into liver biology and disease pathogenesis. Nat. Rev. Gastroenterol. Hepatol. 17, 457–472 (2020).
Lederer, D. J. & Martinez, F. J. Idiopathic pulmonary fibrosis. N. Engl. J. Med. 378, 1811–1823 (2018).
Wolters, P. J., Collard, H. R. & Jones, K. D. Pathogenesis of idiopathic pulmonary fibrosis. Annu Rev. Pathol. 9, 157–179 (2014).
Adams, T. S. et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 6, eaba1983 (2020). scRNA-seq study from human lung fibrosis. A web-based browser of this and other articles is available at http://ipfcellatlas.com
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972 (2020). scRNA-seq study from human lung fibrosis. A web-based browser of this and other articles is available at http://ipfcellatlas.com
Reyfman, P. A. et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med 199, 1517–1536 (2019). scRNA-seq study from human lung fibrosis. A web-based browser of this and other articles is available at http://ipfcellatlas.com
Morse, C. et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 54, 1802441 (2019). scRNA-seq study from human lung fibrosis. A web-based browser of this and other articles is available at http://ipfcellatlas.com
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). Experimental models of lung fibrosis demonstrating the role of monocyte-derived macrophages in the early fibrotic stage after injury.
Wendisch, D. et al. SARS-CoV-2 infection triggers profibrotic macrophage responses and lung fibrosis. Cell 184, 6243–6261 (2021). Analysis of fibrotic changes in COVID-19 lung samples with comparison to IPF.
Wu, X. et al. 3-month, 6-month, 9-month, and 12-month respiratory outcomes in patients following COVID-19-related hospitalisation: a prospective study. Lancet Respir. Med 9, 747–754 (2021).
Faverio, P. et al. One-year pulmonary impairment after severe COVID-19: a prospective, multicenter follow-up study. Respir. Res 23, 65 (2022).
Leslie, J. et al. FPR-1 is an important regulator of neutrophil recruitment and a tissue-specific driver of pulmonary fibrosis. JCI Insight 5, e125937 (2020).
Deng, L., Huang, T. & Zhang, L. T cells in idiopathic pulmonary fibrosis: crucial but controversial. Cell Death Discov. 9, 62 (2023).
Ichikawa, T. et al. CD103hi Treg cells constrain lung fibrosis induced by CD103lo tissue-resident pathogenic CD4 T cells. Nat. Immunol. 20, 1469–1480 (2019).
Chiaramonte, M. G., Donaldson, D. D., Cheever, A. W. & Wynn, T. A. An IL-13 inhibitor blocks the development of hepatic fibrosis during a T-helper type 2-dominated inflammatory response. J. Clin. Invest. 104, 777–785 (1999).
Fallon, P. G., Richardson, E. J., McKenzie, G. J. & McKenzie, A. N. Schistosome infection of transgenic mice defines distinct and contrasting pathogenic roles for IL-4 and IL-13: IL-13 is a profibrotic agent. J. Immunol. 164, 2585–2591 (2000).
Cheever, A. W. et al. Anti-IL-4 treatment of Schistosoma mansoni-infected mice inhibits development of T cells and non-B, non-T cells expressing TH2 cytokines while decreasing egg-induced hepatic fibrosis. J. Immunol. 153, 753–759 (1994).
Raghu, G. et al. SAR156597 in idiopathic pulmonary fibrosis: a phase 2 placebo-controlled study (DRI11772). Eur. Respir. J. 52, 1801130 (2018).
Maher, T. M. et al. Phase 2 trial to assess lebrikizumab in patients with idiopathic pulmonary fibrosis. Eur. Respir. J. 57, 1902442 (2021).
Allanore, Y. et al. A randomised, double-blind, placebo-controlled, 24-week, phase II, proof-of-concept study of romilkimab (SAR156597) in early diffuse cutaneous systemic sclerosis. Ann. Rheum. Dis. 79, 1600–1607 (2020).
Jha, V. et al. Chronic kidney disease: global dimension and perspectives. Lancet 382, 260–272 (2013).
Li, L., Fu, H. & Liu, Y. The fibrogenic niche in kidney fibrosis: components and mechanisms. Nat. Rev. Nephrol. 18, 545–557 (2022).
Kuppe, C. et al. Decoding myofibroblast origins in human kidney fibrosis. Nature 589, 281–286 (2021). scRNA-seq study of healthy versus fibrotic human kidney, generating a cell atlas and highlighting myeloid–mesenchymal interactions.
Hoeft, K. et al. Platelet-instructed SPP1+ macrophages drive myofibroblast activation in fibrosis in a CXCL4-dependent manner. Cell Rep. 42, 112131 (2023). Analysis of human and mouse kidney and cardiac macrophages identifying conserved SAMac populations and demonstrating role of CXCL4 in the generation of SAMacs.
Conway, B. R. et al. Kidney single-cell atlas reveals myeloid heterogeneity in progression and regression of kidney disease. J. Am. Soc. Nephrol. 31, 2833–2854 (2020).
Doke, T. et al. Single-cell analysis identifies the interaction of altered renal tubules with basophils orchestrating kidney fibrosis. Nat. Immunol. 23, 947–959 (2022).
Basile, D. P., Ullah, M. M., Collet, J. A. & Mehrotra, P. T helper 17 cells in the pathophysiology of acute and chronic kidney disease. Kidney Res. Clin. Pract. 40, 12–28 (2021).
Frangogiannis, N. G. Cardiac fibrosis. Cardiovasc Res. 117, 1450–1488 (2021).
Miranda, A. M. A. et al. Single-cell transcriptomics for the assessment of cardiac disease. Nat. Rev. Cardiol. 20, 289–308 (2022).
Koenig, A. L. et al. Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure. Nat. Cardiovasc. Res. 1, 263–280 (2022).
Reichart, D. et al. Pathogenic variants damage cell composition and single cell transcription in cardiomyopathies. Science 377, eabo1984 (2022).
Lavine, K. et al. Targeting immune–fibroblast crosstalk in myocardial infarction and cardiac fibrosis. Preprint at Research Square https://doi.org/10.21203/rs.3.rs-2402606/v1 (2023). Single-cell and spatial analysis of human and mouse cardiac fibrosis, defining disease-associated macrophages and changes in T cells. Identification of IL-1β+ macrophages in the fibrotic niche and demonstration that IL-1β modulation inhibits fibrosis.
Kuppe, C. et al. Spatial multi-omic map of human myocardial infarction. Nature 608, 766–777 (2022). Multiomic analysis of human post-MI hearts, highlighting myeloid–mesenchymal spatial interactions in cardiac repair.
Rao, M. et al. Resolving the intertwining of inflammation and fibrosis in human heart failure at single-cell level. Basic Res. Cardiol. 116, 55 (2021).
Dick, S. A. et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 20, 29–39 (2019).
Bajpai, G. et al. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat. Med. 24, 1234–1245 (2018).
Revelo, X. S. et al. Cardiac resident macrophages prevent fibrosis and stimulate angiogenesis. Circ. Res. 129, 1086–1101 (2021).
Chaffin, M. et al. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy. Nature 608, 174–180 (2022).
Alexanian, M. et al. Chromatin remodeling drives immune–fibroblast crosstalk in heart failure pathogenesis. Preprint at bioRxiv https://doi.org/10.1101/2023.01.06.522937 (2023).
Deniset, J. F. et al. Gata6+ pericardial cavity macrophages relocate to the injured heart and prevent cardiac fibrosis. Immunity 51, 131–140 (2019).
Litvinukova, M. et al. Cells of the adult human heart. Nature 588, 466–472 (2020).
Ong, S. et al. Natural killer cells limit cardiac inflammation and fibrosis by halting eosinophil infiltration. Am. J. Pathol. 185, 847–861 (2015).
Satoh, T. et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 541, 96–101 (2017).
Misharin, A. V. et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J. Exp. Med. 214, 2387–2404 (2017).
Karlmark, K. R. et al. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 50, 261–274 (2009).
Tang, P. M., Nikolic-Paterson, D. J. & Lan, H. Y. Macrophages: versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 15, 144–158 (2019).
Duffield, J. S. et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J. Clin. Invest. 115, 56–65 (2005).
Krenkel, O. et al. Therapeutic inhibition of inflammatory monocyte recruitment reduces steatohepatitis and liver fibrosis. Hepatology 67, 1270–1283 (2018).
Friedman, S. L. et al. A randomized, placebo-controlled trial of cenicriviroc for treatment of nonalcoholic steatohepatitis with fibrosis. Hepatology 67, 1754–1767 (2018).
Buechler, M. B., Fu, W. & Turley, S. J. Fibroblast-macrophage reciprocal interactions in health, fibrosis, and cancer. Immunity 54, 903–915 (2021).
Ding, L. et al. Bone marrow CD11c+ cell-derived amphiregulin promotes pulmonary fibrosis. J. Immunol. 197, 303–312 (2016).
Perugorria, M. J. et al. Non-parenchymal TREM-2 protects the liver from immune-mediated hepatocellular damage. Gut 68, 533–546 (2019).
Labiano, I. et al. TREM-2 plays a protective role in cholestasis by acting as a negative regulator of inflammation. J. Hepatol. 77, 991–1004 (2022).
Li, Z. et al. Single-cell RNA sequencing depicts the local cell landscape in thyroid-associated ophthalmopathy. Cell Rep. Med 3, 100699 (2022).
Raslan, A. A. et al. Single cell transcriptomics of fibrotic lungs unveils aging-associated alterations in endothelial and epithelial cell regeneration. Preprint at bioRxiv https://doi.org/10.1101/2023.01.17.523179 (2023).
Eyres, M. et al. Spatially resolved deconvolution of the fibrotic niche in lung fibrosis. Cell Rep. 40, 111230 (2022).
Chung, B. K., Ogaard, J., Reims, H. M., Karlsen, T. H. & Melum, E. Spatial transcriptomics identifies enriched gene expression and cell types in human liver fibrosis. Hepatol. Commun. 6, 2538–2550 (2022).
Buechler, M. B. et al. Cross-tissue organization of the fibroblast lineage. Nature 593, 575–579 (2021).
Tsukui, T. et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat. Commun. 11, 1920 (2020).
Boyd, D. F. et al. Exuberant fibroblast activity compromises lung function via ADAMTS4. Nature 587, 466–471 (2020).
Filliol, A. et al. Opposing roles of hepatic stellate cell subpopulations in hepatocarcinogenesis. Nature 610, 356–365 (2022).
Tsukui, T. & Sheppard, D. Tracing the origin of pathologic pulmonary fibroblasts. Preprint at bioRxiv https://doi.org/10.1101/2022.11.18.517147 (2022).
Krishnamurty, A. T. et al. LRRC15+ myofibroblasts dictate the stromal setpoint to suppress tumour immunity. Nature 611, 148–154 (2022).
Nguyen, H. N. et al. Autocrine loop involving IL-6 family member LIF, LIF receptor, and STAT4 drives sustained fibroblast production of inflammatory mediators. Immunity 46, 220–232 (2017).
Ng, B. et al. Interleukin-11 is a therapeutic target in idiopathic pulmonary fibrosis. Sci. Transl. Med. 11, eaaw1237 (2019).
Schafer, S. et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 552, 110–115 (2017).
Zhou, X. et al. Circuit design features of a stable two-cell system. Cell 172, 744–757 (2018). This study explains the concept of macrophage–fibroblast circuits and how they regulate cell proliferation and steady-state proportions.
Adler, M. et al. Principles of cell circuits for tissue repair and fibrosis. iScience 23, 100841 (2020). This study explains the concept of macrophage–fibroblast circuits in fibrosis and the idea of ‘hot’ v ‘cold’ fibrosis.
Setten, E. et al. Understanding fibrosis pathogenesis via modeling macrophage-fibroblast interplay in immune-metabolic context. Nat. Commun. 13, 6499 (2022).
Korsunsky, I. et al. Cross-tissue, single-cell stromal atlas identifies shared pathological fibroblast phenotypes in four chronic inflammatory diseases. Med 3, 481–518 (2022).
Wei, K., Nguyen, H. N. & Brenner, M. B. Fibroblast pathology in inflammatory diseases. J. Clin. Invest. 131, e149538 (2021).
Lodyga, M. et al. Cadherin-11-mediated adhesion of macrophages to myofibroblasts establishes a profibrotic niche of active TGF-β. Sci. Signal 12, eaao3469 (2019).
Umetsu, D. T., Katzen, D., Jabara, H. H. & Geha, R. S. Antigen presentation by human dermal fibroblasts: activation of resting T lymphocytes. J. Immunol. 136, 440–445 (1986).
Kundig, T. M. et al. Fibroblasts as efficient antigen-presenting cells in lymphoid organs. Science 268, 1343–1347 (1995).
Elyada, E. et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 9, 1102–1123 (2019).
Kerdidani, D. et al. Lung tumor MHCII immunity depends on in situ antigen presentation by fibroblasts. J. Exp. Med. 219, e20210815 (2022).
Ngwenyama, N. et al. Antigen presentation by cardiac fibroblasts promotes cardiac dysfunction. Nat. Cardiovasc. Res. 1, 761–774 (2022).
Sutherland, T. E., Dyer, D. P. & Allen, J. E. The extracellular matrix and the immune system: a mutually dependent relationship. Science 379, eabp8964 (2023).
Malehmir, M. et al. Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat. Med. 25, 641–655 (2019).
Zhuo, L. et al. SHAP potentiates the CD44-mediated leukocyte adhesion to the hyaluronan substratum. J. Biol. Chem. 281, 20303–20314 (2006).
McQuitty, C. E., Williams, R., Chokshi, S. & Urbani, L. Immunomodulatory role of the extracellular matrix within the liver disease microenvironment. Front. Immunol. 11, 574276 (2020).
Solis, A. G. et al. Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity. Nature 573, 69–74 (2019).
Tharp, K. M. et al. Myeloid mechano-metabolic programming restricts anti-tumor immunity. Preprint at bioRxiv https://doi.org/10.1101/2022.07.14.499764 (2022).
Pakshir, P. et al. Dynamic fibroblast contractions attract remote macrophages in fibrillar collagen matrix. Nat. Commun. 10, 1850 (2019).
Marcellin, P. et al. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a 5-year open-label follow-up study. Lancet 381, 468–475 (2013).
Lassailly, G. et al. Bariatric surgery provides long-term resolution of nonalcoholic steatohepatitis and regression of fibrosis. Gastroenterology 159, 1290–1301 (2020).
Izawa, H. et al. Mineralocorticoid receptor antagonism ameliorates left ventricular diastolic dysfunction and myocardial fibrosis in mildly symptomatic patients with idiopathic dilated cardiomyopathy: a pilot study. Circulation 112, 2940–2945 (2005).
Diez, J. et al. Losartan-dependent regression of myocardial fibrosis is associated with reduction of left ventricular chamber stiffness in hypertensive patients. Circulation 105, 2512–2517 (2002).
Prabhu, S. et al. Regression of diffuse ventricular fibrosis following restoration of sinus rhythm with catheter ablation in patients with atrial fibrillation and systolic dysfunction: a substudy of the CAMERA MRI Trial. JACC Clin. Electrophysiol. 4, 999–1007 (2018).
Fioretto, P., Steffes, M. W., Sutherland, D. E., Goetz, F. C. & Mauer, M. Reversal of lesions of diabetic nephropathy after pancreas transplantation. N. Engl. J. Med. 339, 69–75 (1998).
Fioretto, P., Sutherland, D. E., Najafian, B. & Mauer, M. Remodeling of renal interstitial and tubular lesions in pancreas transplant recipients. Kidney Int. 69, 907–912 (2006).
Ramachandran, P. et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc. Natl Acad. Sci. USA 109, E3186–E3195 (2012). This study demonstrates the role of a subpopulation of matrix-degrading monocyte-derived macrophages in liver fibrosis regression.
Gibbons, M. A. et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am. J. Respir. Crit. Care Med 184, 569–581 (2011).
Rantakari, P. et al. Stabilin-1 expression defines a subset of macrophages that mediate tissue homeostasis and prevent fibrosis in chronic liver injury. Proc. Natl Acad. Sci. USA 113, 9298–9303 (2016).
Takimoto, Y. et al. Myeloid TLR4 signaling promotes post-injury withdrawal resolution of murine liver fibrosis. iScience 26, 106220 (2023).
Pellicoro, A. et al. Elastin accumulation is regulated at the level of degradation by macrophage metalloelastase (MMP-12) during experimental liver fibrosis. Hepatology 55, 1965–1975 (2012).
Fallowfield, J. A. et al. Scar-associated macrophages are a major source of hepatic matrix metalloproteinase-13 and facilitate the resolution of murine hepatic fibrosis. J. Immunol. 178, 5288–5295 (2007).
Ren, J. et al. Twist1 in infiltrating macrophages attenuates kidney fibrosis via matrix metallopeptidase 13-mediated matrix degradation. J. Am. Soc. Nephrol. 30, 1674–1685 (2019).
McKleroy, W., Lee, T. H. & Atabai, K. Always cleave up your mess: targeting collagen degradation to treat tissue fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 304, L709–L721 (2013).
Cui, H. et al. Monocyte-derived alveolar macrophage apolipoprotein E participates in pulmonary fibrosis resolution. JCI Insight 5, e134539 (2020).
Atabai, K. et al. Mfge8 diminishes the severity of tissue fibrosis in mice by binding and targeting collagen for uptake by macrophages. J. Clin. Invest. 119, 3713–3722 (2009).
Madsen, D. H. et al. M2-like macrophages are responsible for collagen degradation through a mannose receptor-mediated pathway. J. Cell Biol. 202, 951–966 (2013).
Campana, L. et al. The STAT3–IL-10–IL-6 pathway is a novel regulator of macrophage efferocytosis and phenotypic conversion in sterile liver injury. J. Immunol. 200, 1169–1187 (2018).
Hu, M. et al. Hepatic macrophages act as a central hub for relaxin-mediated alleviation of liver fibrosis. Nat. Nanotechnol. 16, 466–477 (2021).
Saijou, E. et al. Neutrophils alleviate fibrosis in the CCl4-induced mouse chronic liver injury model. Hepatol. Commun. 2, 703–717 (2018).
Calvente, C. J. et al. Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. J. Clin. Invest. 129, 4091–4109 (2019).
Hegde, P. et al. Mucosal-associated invariant T cells are a profibrogenic immune cell population in the liver. Nat. Commun. 9, 2146 (2018).
Mabire, M. et al. MAIT cell inhibition promotes liver fibrosis regression via macrophage phenotype reprogramming. Nat. Commun. 14, 1830 (2023).
Baeck, C. et al. Pharmacological inhibition of the chemokine C–C motif chemokine ligand 2 (monocyte chemoattractant protein 1) accelerates liver fibrosis regression by suppressing Ly-6C+ macrophage infiltration in mice. Hepatology 59, 1060–1072 (2014).
Kisseleva, T. & Brenner, D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat. Rev. Gastroenterol. Hepatol. 18, 151–166 (2021).
Radaeva, S. et al. Natural killer cells ameliorate liver fibrosis by killing activated stellate cells in NKG2D-dependent and tumor necrosis factor-related apoptosis-inducing ligand-dependent manners. Gastroenterology 130, 435–452 (2006).
Koda, Y. et al. CD8+ tissue-resident memory T cells promote liver fibrosis resolution by inducing apoptosis of hepatic stellate cells. Nat. Commun. 12, 4474 (2021).
Krizhanovsky, V. et al. Senescence of activated stellate cells limits liver fibrosis. Cell 134, 657–667 (2008).
Sagiv, A. et al. Granule exocytosis mediates immune surveillance of senescent cells. Oncogene 32, 1971–1977 (2013).
Troeger, J. S. et al. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology 143, 1073–1083 (2012).
Kisseleva, T. et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc. Natl Acad. Sci. USA 109, 9448–9453 (2012).
Schwantes-An, T. H. et al. Genome-wide association study and meta-analysis on alcohol-associated liver cirrhosis identifies genetic risk factors. Hepatology 73, 1920–1931 (2021).
Anstee, Q. M. et al. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort☆. J. Hepatol. 73, 505–515 (2020).
Allen, R. J. et al. Genome-wide association study of susceptibility to idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med 201, 564–574 (2020).
Nauffal, V. et al. Genetics of myocardial interstitial fibrosis in the human heart and association with disease. Nat. Genet. 55, 777–786 (2023).
Murtha, L. A. et al. The role of pathological aging in cardiac and pulmonary fibrosis. Aging Dis. 10, 419–428 (2019).
Kim, I. H., Kisseleva, T. & Brenner, D. A. Aging and liver disease. Curr. Opin. Gastroenterol. 31, 184–191 (2015).
Lee, S. et al. Molecular programs of fibrotic change in aging human lung. Nat. Commun. 12, 6309 (2021).
Franceschi, C., Garagnani, P., Parini, P., Giuliani, C. & Santoro, A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590 (2018).
Shaw, A. C., Goldstein, D. R. & Montgomery, R. R. Age-dependent dysregulation of innate immunity. Nat. Rev. Immunol. 13, 875–887 (2013).
Gullotta, G. S. et al. Age-induced alterations of granulopoiesis generate atypical neutrophils that aggravate stroke pathology. Nat. Immunol. 24, 925–940 (2023).
Mogilenko, D. A., Shchukina, I. & Artyomov, M. N. Immune ageing at single-cell resolution. Nat. Rev. Immunol. 22, 484–498 (2022).
De Maeyer, R. P. H. & Chambers, E. S. The impact of ageing on monocytes and macrophages. Immunol. Lett. 230, 1–10 (2021).
Govaere, O. et al. A proteo-transcriptomic map of non-alcoholic fatty liver disease signatures. Nat. Metab. 5, 572–578 (2023).
Abozaid, Y. J. et al. Plasma proteomic signature of fatty liver disease: the Rotterdam Study. Hepatology https://doi.org/10.1097/HEP.0000000000000300 (2023).
Sanyal, A. J. et al. Defining the serum proteomic signature of hepatic steatosis, inflammation, ballooning and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 78, 693–703 (2023).
Bowman, W. S. et al. Proteomic biomarkers of progressive fibrosing interstitial lung disease: a multicentre cohort analysis. Lancet Respir. Med 10, 593–602 (2022).
McGlinchey, A. J. et al. Metabolic signatures across the full spectrum of non-alcoholic fatty liver disease. JHEP Rep. 4, 100477 (2022).
Seeliger, B. et al. Changes in serum metabolomics in idiopathic pulmonary fibrosis and effect of approved antifibrotic medication. Front Pharm. 13, 837680 (2022).
Sacchi, M., Bansal, R. & Rouwkema, J. Bioengineered 3D models to recapitulate tissue fibrosis. Trends Biotechnol. 38, 623–636 (2020).
Acknowledgements
M.B. is supported by a US DOD CDMRP Investigator-Initiated Research Award (W81XWH2110417). P.R. is supported by a Medical Research Council Senior Clinical Fellowship (MR/W015919/1). Images were made with copyright (M.B.) using Biorender.
Author information
Authors and Affiliations
Contributions
M.B. and P.R. conceived and co-wrote the manuscript, with equal contribution.
Corresponding authors
Ethics declarations
Competing interests
M.B has performed consultancy work for Merck and Ono Pharma. P.R. has received research support from Genentech, Intercept Pharmaceuticals, and NeoGenomics. P.R. has performed consultancy work for Merck.
Peer review
Peer review information
Nature Immunology thanks Thomas Wynn and Matthew Buechler for their contribution to the peer review of this work. Jamie D. K. Wilson, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhattacharya, M., Ramachandran, P. Immunology of human fibrosis. Nat Immunol 24, 1423–1433 (2023). https://doi.org/10.1038/s41590-023-01551-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-023-01551-9